Machine Learning Results (Mod11)
Fitting three machine learning models using the TidyModel Framework.
NOTE: Original code for exercise was divided into separate scripts in respective repo.
## OBJECTIVE
Data
Loading Data
Loading all the default settings and preliminary programs.
Path to Processed Data and loading of cleaned data
data_location <- here::here("data","processed_data","processeddata.rds")
data<- readRDS(data_location)Reminder: Outcome of interest is Body Temp; Categorical outcome is Nausea; Predictor= RunnyNose
glimpse(data)## Rows: 730
## Columns: 32
## $ SwollenLymphNodes <fct> Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, No, Yes, Y~
## $ ChestCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
## $ ChillsSweats <fct> No, No, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, ~
## $ NasalCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
## $ CoughYN <fct> Yes, Yes, No, Yes, No, Yes, Yes, Yes, Yes, Yes, No, ~
## $ Sneeze <fct> No, No, Yes, Yes, No, Yes, No, Yes, No, No, No, No, ~
## $ Fatigue <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
## $ SubjectiveFever <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes~
## $ Headache <fct> Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes~
## $ Weakness <fct> Mild, Severe, Severe, Severe, Moderate, Moderate, Mi~
## $ WeaknessYN <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
## $ CoughIntensity <fct> Severe, Severe, Mild, Moderate, None, Moderate, Seve~
## $ CoughYN2 <fct> Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes, Yes, Yes~
## $ Myalgia <fct> Mild, Severe, Severe, Severe, Mild, Moderate, Mild, ~
## $ MyalgiaYN <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
## $ RunnyNose <fct> No, No, Yes, Yes, No, No, Yes, Yes, Yes, Yes, No, No~
## $ AbPain <fct> No, No, Yes, No, No, No, No, No, No, No, Yes, Yes, N~
## $ ChestPain <fct> No, No, Yes, No, No, Yes, Yes, No, No, No, No, Yes, ~
## $ Diarrhea <fct> No, No, No, No, No, Yes, No, No, No, No, No, No, No,~
## $ EyePn <fct> No, No, No, No, Yes, No, No, No, No, No, Yes, No, Ye~
## $ Insomnia <fct> No, No, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Yes, Y~
## $ ItchyEye <fct> No, No, No, No, No, No, No, No, No, No, No, No, Yes,~
## $ Nausea <fct> No, No, Yes, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Y~
## $ EarPn <fct> No, Yes, No, Yes, No, No, No, No, No, No, No, Yes, Y~
## $ Hearing <fct> No, Yes, No, No, No, No, No, No, No, No, No, No, No,~
## $ Pharyngitis <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, ~
## $ Breathless <fct> No, No, Yes, No, No, Yes, No, No, No, Yes, No, Yes, ~
## $ ToothPn <fct> No, No, Yes, No, No, No, No, No, Yes, No, No, Yes, N~
## $ Vision <fct> No, No, No, No, No, No, No, No, No, No, No, No, No, ~
## $ Vomit <fct> No, No, No, No, No, No, Yes, No, No, No, Yes, Yes, N~
## $ Wheeze <fct> No, No, No, Yes, No, Yes, No, No, No, No, No, Yes, N~
## $ BodyTemp <dbl> 98.3, 100.4, 100.8, 98.8, 100.5, 98.4, 102.5, 98.4, ~Feature / Variable removal
- Remove severity score data
OrderedData<-data%>%
#remove YN variables for those variables with severity factors
select(-WeaknessYN,-CoughYN,-MyalgiaYN,-CoughYN2)%>%
#code symptom severity factors as ordinal
mutate(Weakness=as.ordered(Weakness), CoughIntensity=as.ordered(CoughIntensity), Myalgia=as.ordered(Myalgia))%>%
#Order severity in ordered factors: None<mild<Moderate<Severee
mutate_at(vars(Weakness, CoughIntensity, Myalgia),
list(~factor(.,levels = c("None","Mild","Moderate","Severe"),ordered = TRUE)))
summary(OrderedData)## SwollenLymphNodes ChestCongestion ChillsSweats NasalCongestion Sneeze
## No :418 No :323 No :130 No :167 No :339
## Yes:312 Yes:407 Yes:600 Yes:563 Yes:391
##
##
##
##
## Fatigue SubjectiveFever Headache Weakness CoughIntensity
## No : 64 No :230 No :115 None : 49 None : 47
## Yes:666 Yes:500 Yes:615 Mild :223 Mild :154
## Moderate:338 Moderate:357
## Severe :120 Severe :172
##
##
## Myalgia RunnyNose AbPain ChestPain Diarrhea EyePn Insomnia
## None : 79 No :211 No :639 No :497 No :631 No :617 No :315
## Mild :213 Yes:519 Yes: 91 Yes:233 Yes: 99 Yes:113 Yes:415
## Moderate:325
## Severe :113
##
##
## ItchyEye Nausea EarPn Hearing Pharyngitis Breathless ToothPn
## No :551 No :475 No :568 No :700 No :119 No :436 No :565
## Yes:179 Yes:255 Yes:162 Yes: 30 Yes:611 Yes:294 Yes:165
##
##
##
##
## Vision Vomit Wheeze BodyTemp
## No :711 No :652 No :510 Min. : 97.20
## Yes: 19 Yes: 78 Yes:220 1st Qu.: 98.20
## Median : 98.50
## Mean : 98.94
## 3rd Qu.: 99.30
## Max. :103.10Low variance predictors view summary of Ordered Data <50 entries
Mod11Data<-OrderedData%>%
select(-Hearing, -Vision)
glimpse(Mod11Data) ## Rows: 730
## Columns: 26
## $ SwollenLymphNodes <fct> Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, No, Yes, Y~
## $ ChestCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
## $ ChillsSweats <fct> No, No, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, ~
## $ NasalCongestion <fct> No, Yes, Yes, Yes, No, No, No, Yes, Yes, Yes, Yes, Y~
## $ Sneeze <fct> No, No, Yes, Yes, No, Yes, No, Yes, No, No, No, No, ~
## $ Fatigue <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Ye~
## $ SubjectiveFever <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, Yes~
## $ Headache <fct> Yes, Yes, Yes, Yes, Yes, Yes, No, Yes, Yes, Yes, Yes~
## $ Weakness <ord> Mild, Severe, Severe, Severe, Moderate, Moderate, Mi~
## $ CoughIntensity <ord> Severe, Severe, Mild, Moderate, None, Moderate, Seve~
## $ Myalgia <ord> Mild, Severe, Severe, Severe, Mild, Moderate, Mild, ~
## $ RunnyNose <fct> No, No, Yes, Yes, No, No, Yes, Yes, Yes, Yes, No, No~
## $ AbPain <fct> No, No, Yes, No, No, No, No, No, No, No, Yes, Yes, N~
## $ ChestPain <fct> No, No, Yes, No, No, Yes, Yes, No, No, No, No, Yes, ~
## $ Diarrhea <fct> No, No, No, No, No, Yes, No, No, No, No, No, No, No,~
## $ EyePn <fct> No, No, No, No, Yes, No, No, No, No, No, Yes, No, Ye~
## $ Insomnia <fct> No, No, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Yes, Y~
## $ ItchyEye <fct> No, No, No, No, No, No, No, No, No, No, No, No, Yes,~
## $ Nausea <fct> No, No, Yes, Yes, Yes, Yes, No, No, Yes, Yes, Yes, Y~
## $ EarPn <fct> No, Yes, No, Yes, No, No, No, No, No, No, No, Yes, Y~
## $ Pharyngitis <fct> Yes, Yes, Yes, Yes, Yes, Yes, Yes, No, No, No, Yes, ~
## $ Breathless <fct> No, No, Yes, No, No, Yes, No, No, No, Yes, No, Yes, ~
## $ ToothPn <fct> No, No, Yes, No, No, No, No, No, Yes, No, No, Yes, N~
## $ Vomit <fct> No, No, No, No, No, No, Yes, No, No, No, Yes, Yes, N~
## $ Wheeze <fct> No, No, No, Yes, No, Yes, No, No, No, No, No, Yes, N~
## $ BodyTemp <dbl> 98.3, 100.4, 100.8, 98.8, 100.5, 98.4, 102.5, 98.4, ~location to save file
save_data_location <- here::here("data","processed_data","Mod11Data.rds")
saveRDS(Mod11Data, file = save_data_location)Summary of data
skim(Mod11Data) # use skimmer to summarize data| Name | Mod11Data |
| Number of rows | 730 |
| Number of columns | 26 |
| _______________________ | |
| Column type frequency: | |
| factor | 25 |
| numeric | 1 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| SwollenLymphNodes | 0 | 1 | FALSE | 2 | No: 418, Yes: 312 |
| ChestCongestion | 0 | 1 | FALSE | 2 | Yes: 407, No: 323 |
| ChillsSweats | 0 | 1 | FALSE | 2 | Yes: 600, No: 130 |
| NasalCongestion | 0 | 1 | FALSE | 2 | Yes: 563, No: 167 |
| Sneeze | 0 | 1 | FALSE | 2 | Yes: 391, No: 339 |
| Fatigue | 0 | 1 | FALSE | 2 | Yes: 666, No: 64 |
| SubjectiveFever | 0 | 1 | FALSE | 2 | Yes: 500, No: 230 |
| Headache | 0 | 1 | FALSE | 2 | Yes: 615, No: 115 |
| Weakness | 0 | 1 | TRUE | 4 | Mod: 338, Mil: 223, Sev: 120, Non: 49 |
| CoughIntensity | 0 | 1 | TRUE | 4 | Mod: 357, Sev: 172, Mil: 154, Non: 47 |
| Myalgia | 0 | 1 | TRUE | 4 | Mod: 325, Mil: 213, Sev: 113, Non: 79 |
| RunnyNose | 0 | 1 | FALSE | 2 | Yes: 519, No: 211 |
| AbPain | 0 | 1 | FALSE | 2 | No: 639, Yes: 91 |
| ChestPain | 0 | 1 | FALSE | 2 | No: 497, Yes: 233 |
| Diarrhea | 0 | 1 | FALSE | 2 | No: 631, Yes: 99 |
| EyePn | 0 | 1 | FALSE | 2 | No: 617, Yes: 113 |
| Insomnia | 0 | 1 | FALSE | 2 | Yes: 415, No: 315 |
| ItchyEye | 0 | 1 | FALSE | 2 | No: 551, Yes: 179 |
| Nausea | 0 | 1 | FALSE | 2 | No: 475, Yes: 255 |
| EarPn | 0 | 1 | FALSE | 2 | No: 568, Yes: 162 |
| Pharyngitis | 0 | 1 | FALSE | 2 | Yes: 611, No: 119 |
| Breathless | 0 | 1 | FALSE | 2 | No: 436, Yes: 294 |
| ToothPn | 0 | 1 | FALSE | 2 | No: 565, Yes: 165 |
| Vomit | 0 | 1 | FALSE | 2 | No: 652, Yes: 78 |
| Wheeze | 0 | 1 | FALSE | 2 | No: 510, Yes: 220 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| BodyTemp | 0 | 1 | 98.94 | 1.2 | 97.2 | 98.2 | 98.5 | 99.3 | 103.1 | ▇▇▂▁▁ |
Set Seed
#set random seed to 123 for reproducibility
set.seed(123)Training and Testing Split
#split dataset into 70% training, 30% testing
#use BodyTemp as stratification
data_split <- initial_split(Mod11Data,
prop = 7/10,
strata = BodyTemp)
#create dataframes for the split data
train_data <- training(data_split)
test_data <- testing(data_split)5-fold Cross Validation
folds <-
vfold_cv(train_data,
v = 5,
repeats = 5,
strata = BodyTemp)Body Temperature vs. all predictors
Below I am creating the new full model recipe for body temperature against all predictors, We have the recipe program add dummy values for all nominal predictors– BodyTemp
Mod11_rec<-
recipe(BodyTemp~., train_data)%>%
step_dummy(all_nominal_predictors())Null Model Perfomance
A null model is one with out any predictors. In this case, this predicts the mean of the outcome. We will compute the RMSE for this and compare with the final model.
Create Null Model
#create null model
null_mod <-
null_model() %>%
set_engine("parsnip") %>%
set_mode("regression")
#add recipe and model into workflow
null_wflow <-
workflow() %>%
add_recipe(Mod11_rec) %>%
add_model(null_mod)Fitting the null model to training data
null_train <-
null_wflow %>%
fit(data = train_data)
#summary of null model with training data to get mean (which in this case is the RMSE)
tidy(null_train)## # A tibble: 1 x 1
## value
## <dbl>
## 1 98.9Fitting the null model to test data
null_test <-
null_wflow %>%
fit(data = test_data)
#summary of null model with test data to get mean (which in this case is the RMSE)
tidy(null_train)## # A tibble: 1 x 1
## value
## <dbl>
## 1 98.9Null RMSE values
#RMSE for training data
null_RMSE_train <-
tibble(
rmse = rmse_vec(
truth = train_data$BodyTemp,
estimate = rep(mean(train_data$BodyTemp),
nrow(train_data))),
SE = 0,
model = "Null - Train")
null_RMSE_train## # A tibble: 1 x 3
## rmse SE model
## <dbl> <dbl> <chr>
## 1 1.21 0 Null - Train#RMSE for testing data
null_RMSE_test <-
tibble(
rmse = rmse_vec(
truth = test_data$BodyTemp,
estimate = rep(mean(test_data$BodyTemp),
nrow(test_data))),
SE = 0,
model = "Null - Test")
null_RMSE_test## # A tibble: 1 x 3
## rmse SE model
## <dbl> <dbl> <chr>
## 1 1.16 0 Null - TestDecision Tree Model
AKA, Tree Model, it is a non-parametric supervised learning method used for classification and regression. The goal is to create a model that predicts the value of a target variable by learning simple decision rules inferred from the data features. A tree can be seen as a piecewise constant approximation.
Structure of this code is biased from TidyModels Tutorial for Tuning.
Model Evaluation
Defining the tree model.
tree_mod<-
#parsnip package - tuning hyperparameters-- with set engine and mode
decision_tree(
# tune() is a placeholder for parsnip to ID
cost_complexity = tune(),
tree_depth = tune(),
min_n = tune())%>%
set_engine("rpart")%>%
set_mode("regression") #use regression instead of classification
tree_mod## Decision Tree Model Specification (regression)
##
## Main Arguments:
## cost_complexity = tune()
## tree_depth = tune()
## min_n = tune()
##
## Computational engine: rpartTuning grid specification
tree_grid<-
# grid_regular- chooses sensible values for each hyper parameter
grid_regular(
cost_complexity(),
tree_depth(),
min_n(), #add to increase 25x
levels = 5
)
tree_grid## # A tibble: 125 x 3
## cost_complexity tree_depth min_n
## <dbl> <int> <int>
## 1 0.0000000001 1 2
## 2 0.0000000178 1 2
## 3 0.00000316 1 2
## 4 0.000562 1 2
## 5 0.1 1 2
## 6 0.0000000001 4 2
## 7 0.0000000178 4 2
## 8 0.00000316 4 2
## 9 0.000562 4 2
## 10 0.1 4 2
## # ... with 115 more rows# view the tree depth
tree_grid%>%
count(tree_depth)## # A tibble: 5 x 2
## tree_depth n
## <int> <int>
## 1 1 25
## 2 4 25
## 3 8 25
## 4 11 25
## 5 15 25Define workflow
tree_WF<-
workflow()%>%
add_model(tree_mod)%>% # Preprocessor, decision tree and added dummy
add_recipe(Mod11_rec) # Model- recipe from `Recipe 1` chunk, features BodyTemp and all other predictors
tree_WF## == Workflow ====================================================================
## Preprocessor: Recipe
## Model: decision_tree()
##
## -- Preprocessor ----------------------------------------------------------------
## 1 Recipe Step
##
## * step_dummy()
##
## -- Model -----------------------------------------------------------------------
## Decision Tree Model Specification (regression)
##
## Main Arguments:
## cost_complexity = tune()
## tree_depth = tune()
## min_n = tune()
##
## Computational engine: rpartCross-Validation and tune_grid() for additional tuning
#tune the model with previously specified CV and RMSE
tree_res <-
tree_WF %>%
tune_grid(
resamples=folds,
grid= tree_grid,
metric_set(rmse))## ! Fold1, Repeat1: internal: A correlation computation is required, but `estimate` is const...## ! Fold2, Repeat1: internal: A correlation computation is required, but `estimate` is const...## ! Fold3, Repeat1: internal: A correlation computation is required, but `estimate` is const...## ! Fold4, Repeat1: internal: A correlation computation is required, but `estimate` is const...## ! Fold5, Repeat1: internal: A correlation computation is required, but `estimate` is const...## ! Fold1, Repeat2: internal: A correlation computation is required, but `estimate` is const...## ! Fold2, Repeat2: internal: A correlation computation is required, but `estimate` is const...## ! Fold3, Repeat2: internal: A correlation computation is required, but `estimate` is const...## ! Fold4, Repeat2: internal: A correlation computation is required, but `estimate` is const...## ! Fold5, Repeat2: internal: A correlation computation is required, but `estimate` is const...## ! Fold1, Repeat3: internal: A correlation computation is required, but `estimate` is const...## ! Fold2, Repeat3: internal: A correlation computation is required, but `estimate` is const...## ! Fold3, Repeat3: internal: A correlation computation is required, but `estimate` is const...## ! Fold4, Repeat3: internal: A correlation computation is required, but `estimate` is const...## ! Fold5, Repeat3: internal: A correlation computation is required, but `estimate` is const...## ! Fold1, Repeat4: internal: A correlation computation is required, but `estimate` is const...## ! Fold2, Repeat4: internal: A correlation computation is required, but `estimate` is const...## ! Fold3, Repeat4: internal: A correlation computation is required, but `estimate` is const...## ! Fold4, Repeat4: internal: A correlation computation is required, but `estimate` is const...## ! Fold5, Repeat4: internal: A correlation computation is required, but `estimate` is const...## ! Fold1, Repeat5: internal: A correlation computation is required, but `estimate` is const...## ! Fold2, Repeat5: internal: A correlation computation is required, but `estimate` is const...## ! Fold3, Repeat5: internal: A correlation computation is required, but `estimate` is const...## ! Fold4, Repeat5: internal: A correlation computation is required, but `estimate` is const...## ! Fold5, Repeat5: internal: A correlation computation is required, but `estimate` is const...Decision Tree Metrics and Plotting - Model Evaluation
autoplot
creates a defaulted visualization
tree_res%>%
autoplot()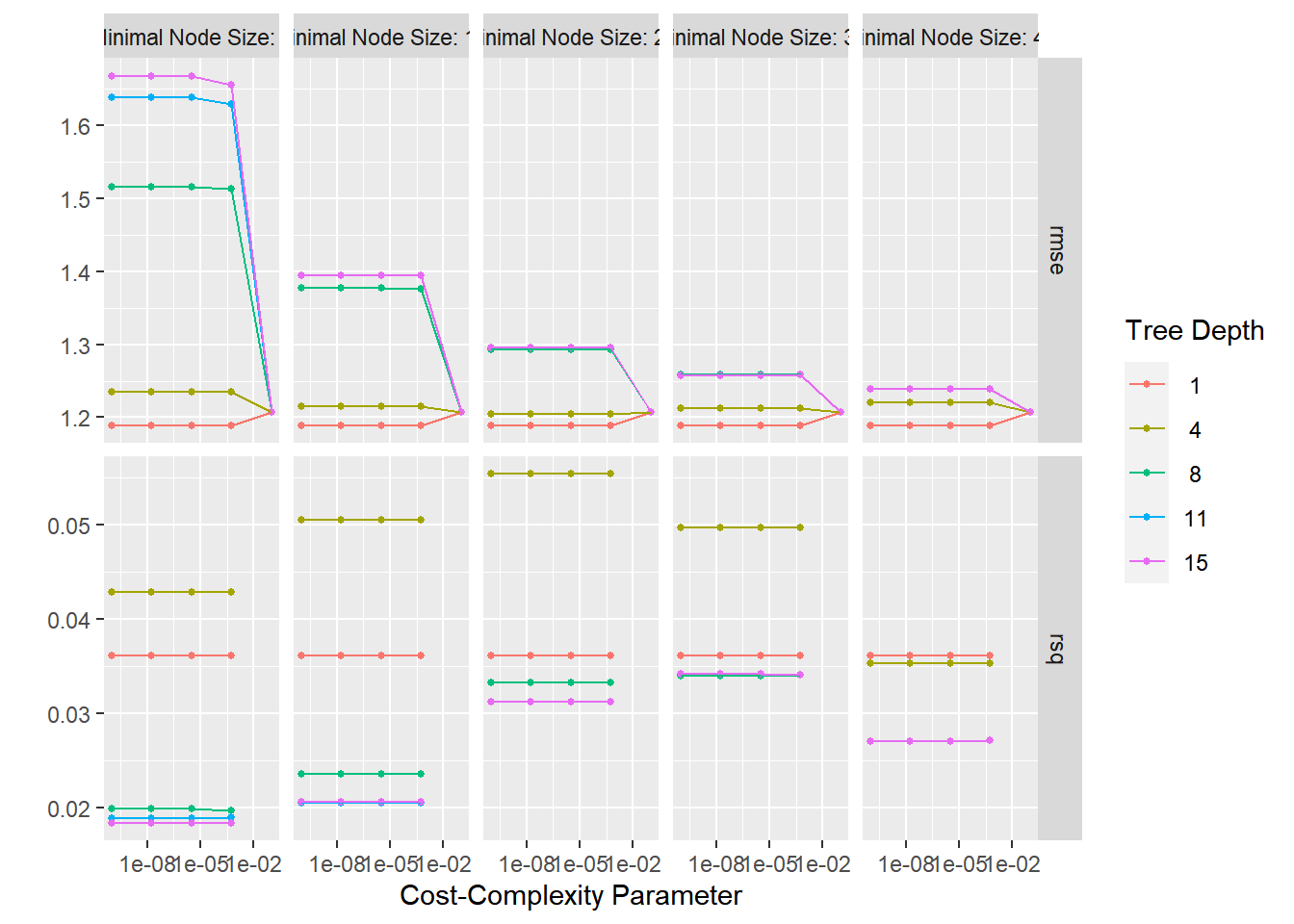
Finding the Best Tree Model
best_tree <-
tree_res %>%
select_best("rmse") #function to pull out the single set of hyperparameter values for best decision tree model
best_tree## # A tibble: 1 x 4
## cost_complexity tree_depth min_n .config
## <dbl> <int> <int> <chr>
## 1 0.0000000001 1 2 Preprocessor1_Model001# results for tree depth and cost complexity that max the accuracy in the dataset of cell images.Finalize Decision Tree
Final Workflow
final_WF <-
tree_WF %>%
finalize_workflow(best_tree) # already pulled best "RMSE" tree
final_WF## == Workflow ====================================================================
## Preprocessor: Recipe
## Model: decision_tree()
##
## -- Preprocessor ----------------------------------------------------------------
## 1 Recipe Step
##
## * step_dummy()
##
## -- Model -----------------------------------------------------------------------
## Decision Tree Model Specification (regression)
##
## Main Arguments:
## cost_complexity = 1e-10
## tree_depth = 1
## min_n = 2
##
## Computational engine: rpartLast Fit
Fitting with fit()
final_fit<-
final_WF%>%
fit(train_data)Predicting outcomes for final model (training data)
tree_pred<-
predict(final_fit,
train_data) #testing fit with the training splitPlotting final tree
rpart.plot(extract_fit_parsnip(final_fit)$fit)## Warning: Cannot retrieve the data used to build the model (model.frame: object '..y' not found).
## To silence this warning:
## Call rpart.plot with roundint=FALSE,
## or rebuild the rpart model with model=TRUE.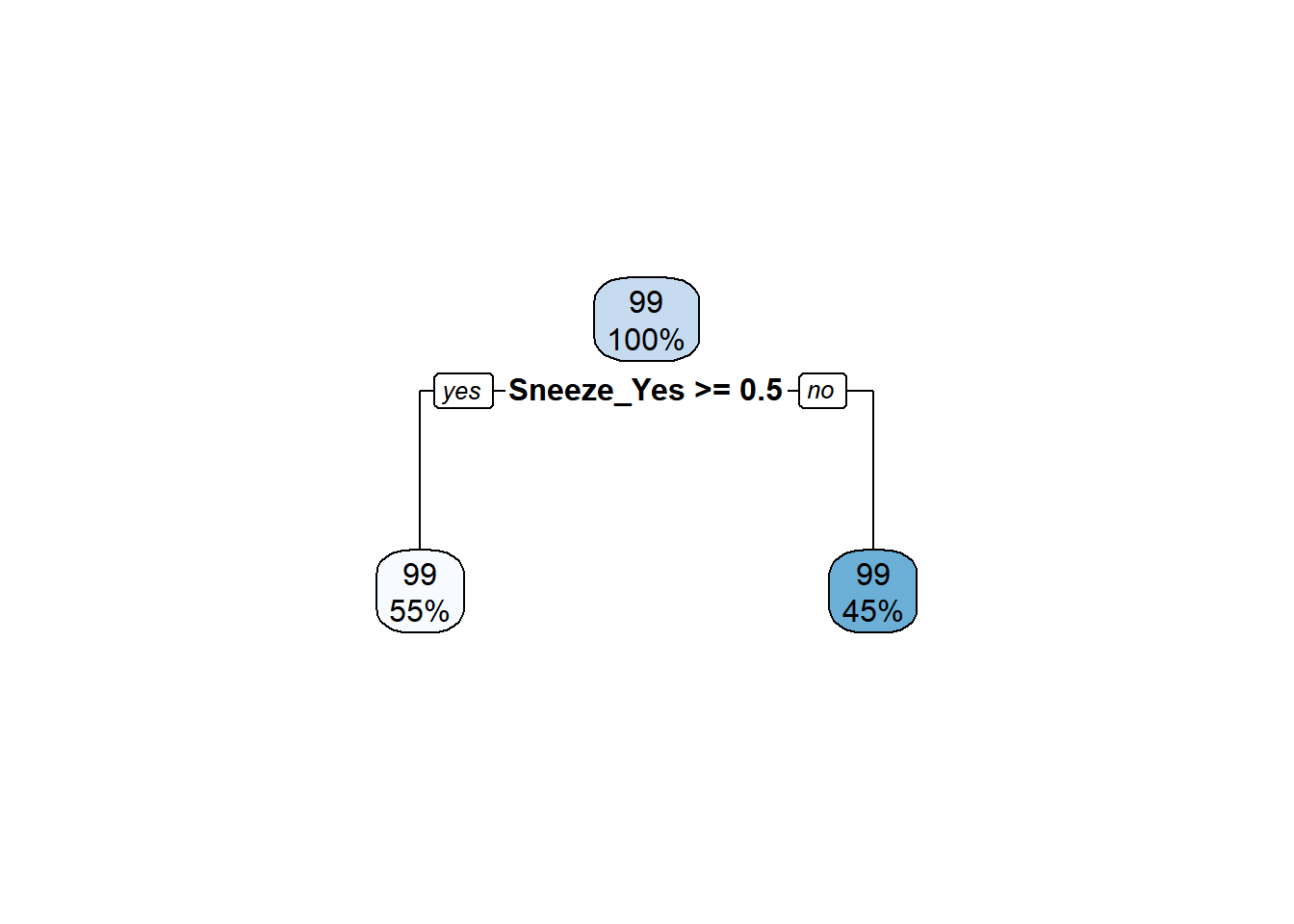
Plotting
Predictions and Interval Fit
#predicted versus observed
plot(tree_pred$.pred,train_data$BodyTemp)
abline(a=0,b=1, col = 'red') #45 degree line, along which the results should fall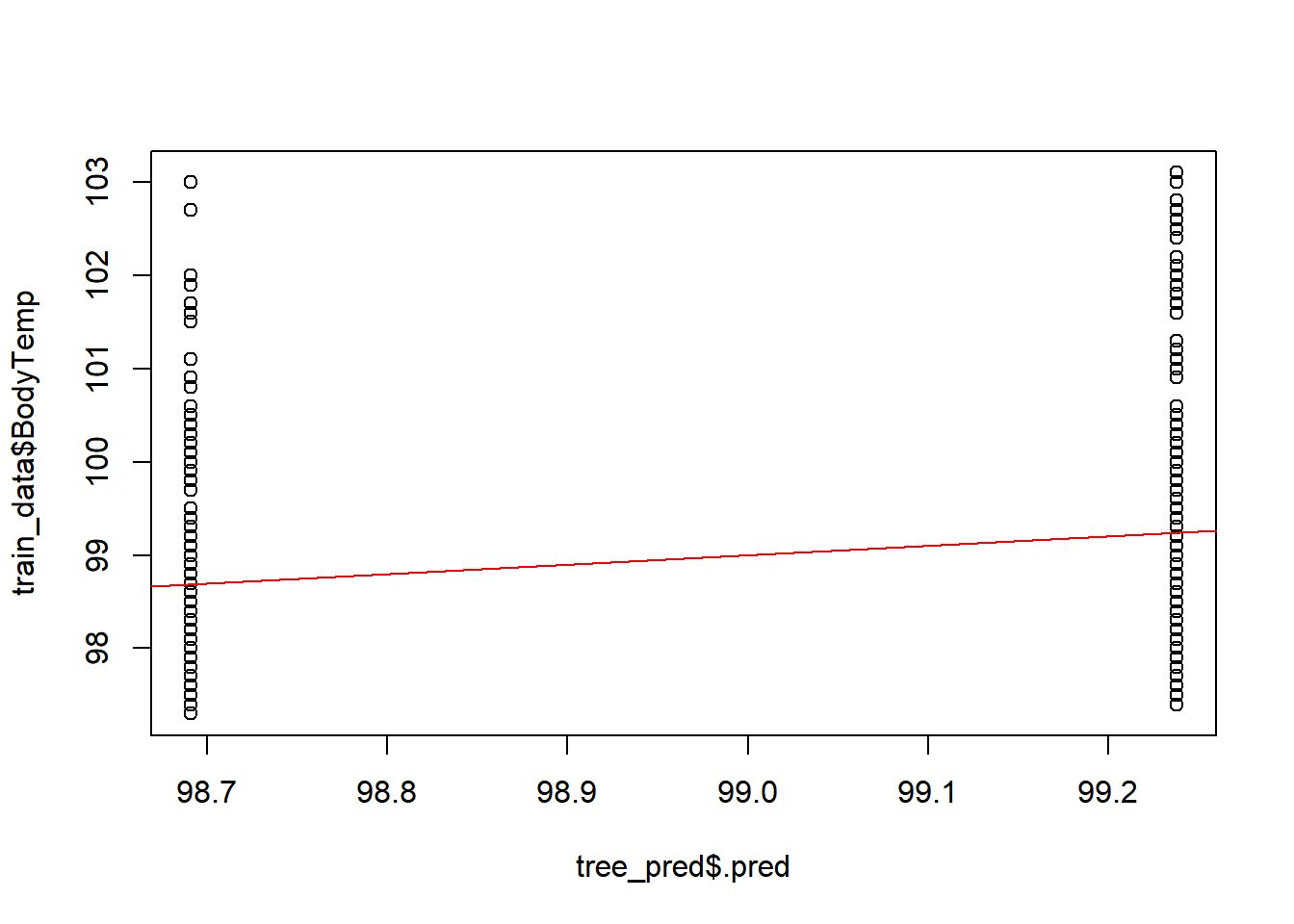 ### Residuals
### Residuals
plot(tree_pred$.pred-train_data$BodyTemp)
abline(a=0,b=0, col = 'red') #straight line, along which the results should fall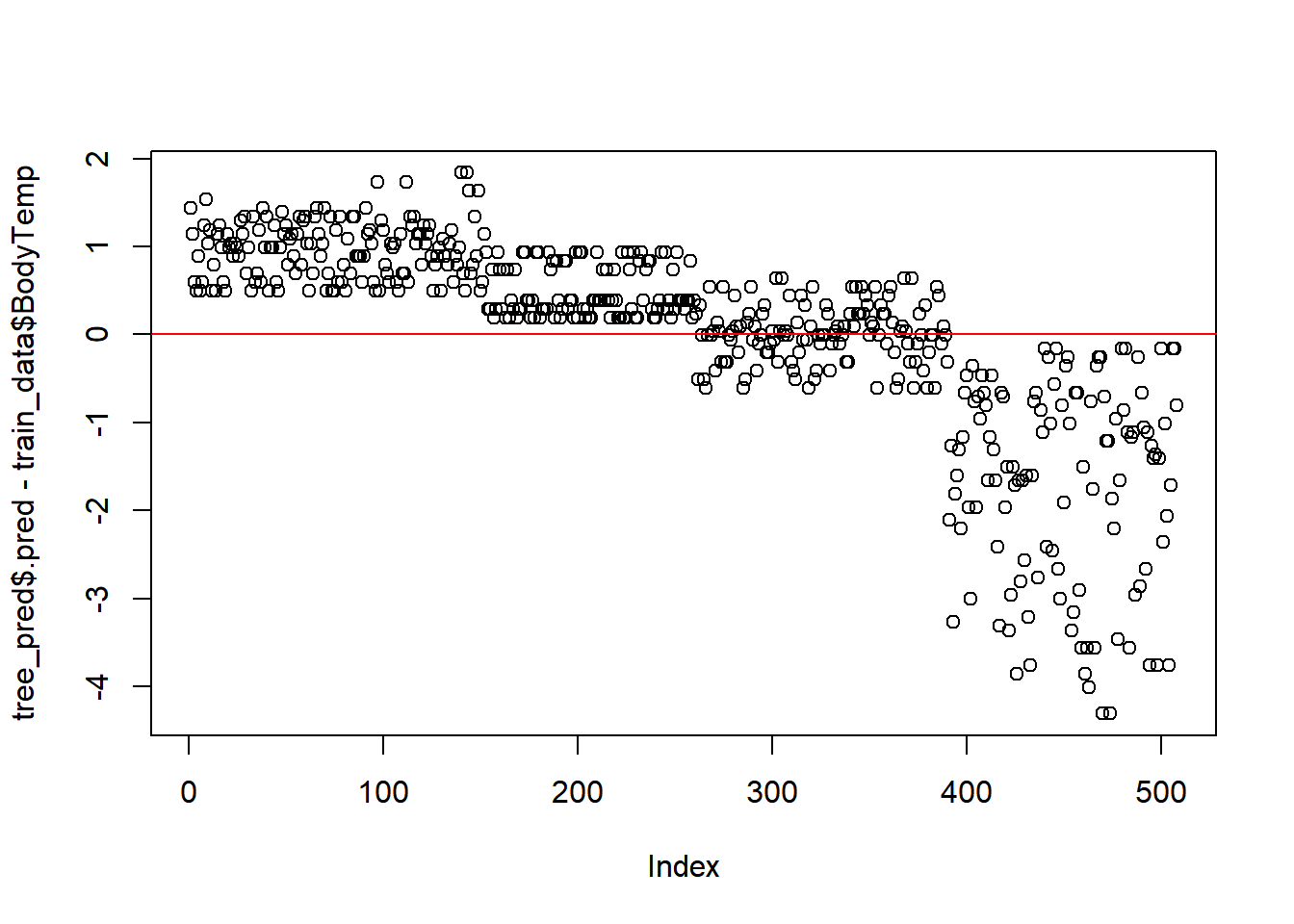
Tree Performance
tree_perfomance <- tree_res %>% show_best(n = 5)## Warning: No value of `metric` was given; metric 'rmse' will be used.print(tree_perfomance)## # A tibble: 5 x 9
## cost_complexity tree_depth min_n .metric .estimator mean n std_err
## <dbl> <int> <int> <chr> <chr> <dbl> <int> <dbl>
## 1 0.0000000001 1 2 rmse standard 1.19 25 0.0181
## 2 0.0000000178 1 2 rmse standard 1.19 25 0.0181
## 3 0.00000316 1 2 rmse standard 1.19 25 0.0181
## 4 0.000562 1 2 rmse standard 1.19 25 0.0181
## 5 0.0000000001 1 11 rmse standard 1.19 25 0.0181
## # ... with 1 more variable: .config <chr>Compare model performance to null model
tree_RMSE<-
tree_res%>% #CV and tuned grid recipe
show_best(n=1)%>%
transmute( # row names in the performance output
rmse=round(mean,2),
SE=round(std_err,2),
model="Tree")## Warning: No value of `metric` was given; metric 'rmse' will be used.tree_RMSE## # A tibble: 1 x 3
## rmse SE model
## <dbl> <dbl> <chr>
## 1 1.19 0.02 TreeComments:
The best performing tree model predicts two values.
LASSO for Linear Regression
Type of linear regression that uses shrinkage (data values shrunk towards a central point, mean).
NOTE: Aspects from the Decision Tree Model will be used to perform a LASSO Linear Regression. Code for this section may be taken from (TidyModels Tutorial Case Study)[https://www.tidymodels.org/start/case-study/].
LASSO Setup
Building/define the LASSO model
lasso_model <- linear_reg() %>%
set_mode("regression") %>%
set_engine("glmnet") %>% #glmnet engine to specify a penelized logistic regression model
set_args(penalty = tune(),
mixture = 1) #mixture 1 => means we use the LASSO modelLASSO WorkFlow
lasso_WF<-
workflow()%>%
add_model(lasso_model)%>%
add_recipe(Mod11_rec)LASSO Tuning
#specifics for tuning grid = add penalties
lasso_grid <-
tibble(penalty = 10^seq(-3, 0, length.out = 30))#tune model
lasso_tune_rec <-
lasso_WF %>%
tune_grid(resamples = folds, #tune with CV value pre designated
grid = lasso_grid,
control = control_grid(
save_pred = TRUE
),
metrics = metric_set(rmse) #RMSE as target metric
)lasso_tune_rec%>%
autoplot()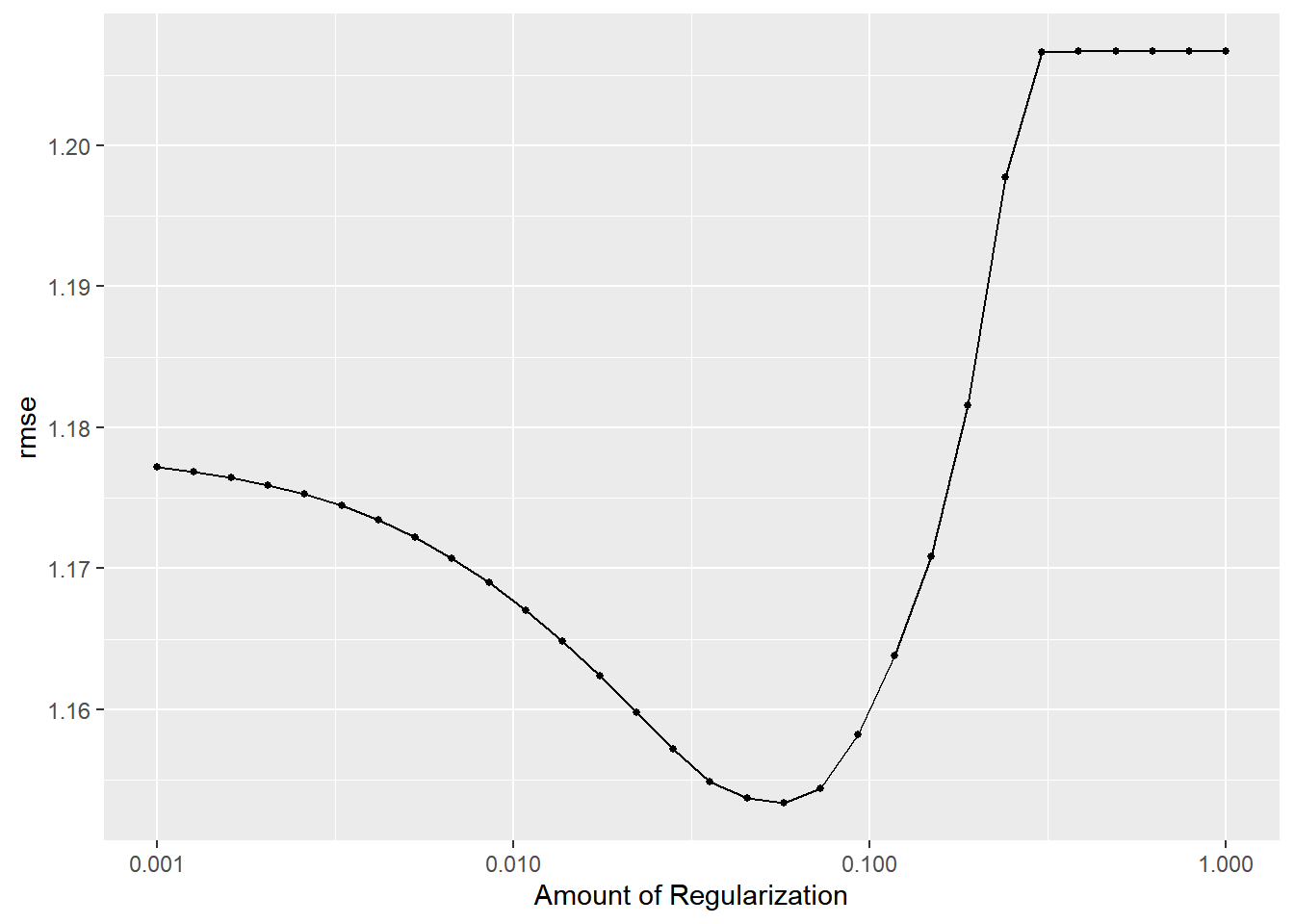
#View top 15 models with lowest RMSEs
lasso_top_models<-
lasso_tune_rec%>%
show_best("rmse", n=15)%>%
arrange(penalty)
lasso_top_models## # A tibble: 15 x 7
## penalty .metric .estimator mean n std_err .config
## <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 0.00530 rmse standard 1.17 25 0.0167 Preprocessor1_Model08
## 2 0.00672 rmse standard 1.17 25 0.0167 Preprocessor1_Model09
## 3 0.00853 rmse standard 1.17 25 0.0167 Preprocessor1_Model10
## 4 0.0108 rmse standard 1.17 25 0.0167 Preprocessor1_Model11
## 5 0.0137 rmse standard 1.16 25 0.0167 Preprocessor1_Model12
## 6 0.0174 rmse standard 1.16 25 0.0167 Preprocessor1_Model13
## 7 0.0221 rmse standard 1.16 25 0.0168 Preprocessor1_Model14
## 8 0.0281 rmse standard 1.16 25 0.0169 Preprocessor1_Model15
## 9 0.0356 rmse standard 1.15 25 0.0169 Preprocessor1_Model16
## 10 0.0452 rmse standard 1.15 25 0.0169 Preprocessor1_Model17
## 11 0.0574 rmse standard 1.15 25 0.0169 Preprocessor1_Model18
## 12 0.0728 rmse standard 1.15 25 0.0170 Preprocessor1_Model19
## 13 0.0924 rmse standard 1.16 25 0.0172 Preprocessor1_Model20
## 14 0.117 rmse standard 1.16 25 0.0175 Preprocessor1_Model21
## 15 0.149 rmse standard 1.17 25 0.0178 Preprocessor1_Model22#Best tuned LASSO model
lasso_best<-
lasso_tune_rec%>%
select_best(metric="rmse")
# finalize workflow with best model
lasso_best_WF <-
lasso_WF %>%
finalize_workflow(lasso_best)
# fitting best performing model
lasso_best_fit <-
lasso_best_WF %>%
fit(data = train_data)
lasso_pred <-
predict(lasso_best_fit, train_data)LASSO Evaluation
Tuning parameters plot in LASSO
# extract models from the final fit
x<-
lasso_best_fit$fit$fit$fit
# plotting the number of predictors and thier changes with the tuning parameter
plot(x, "lambda")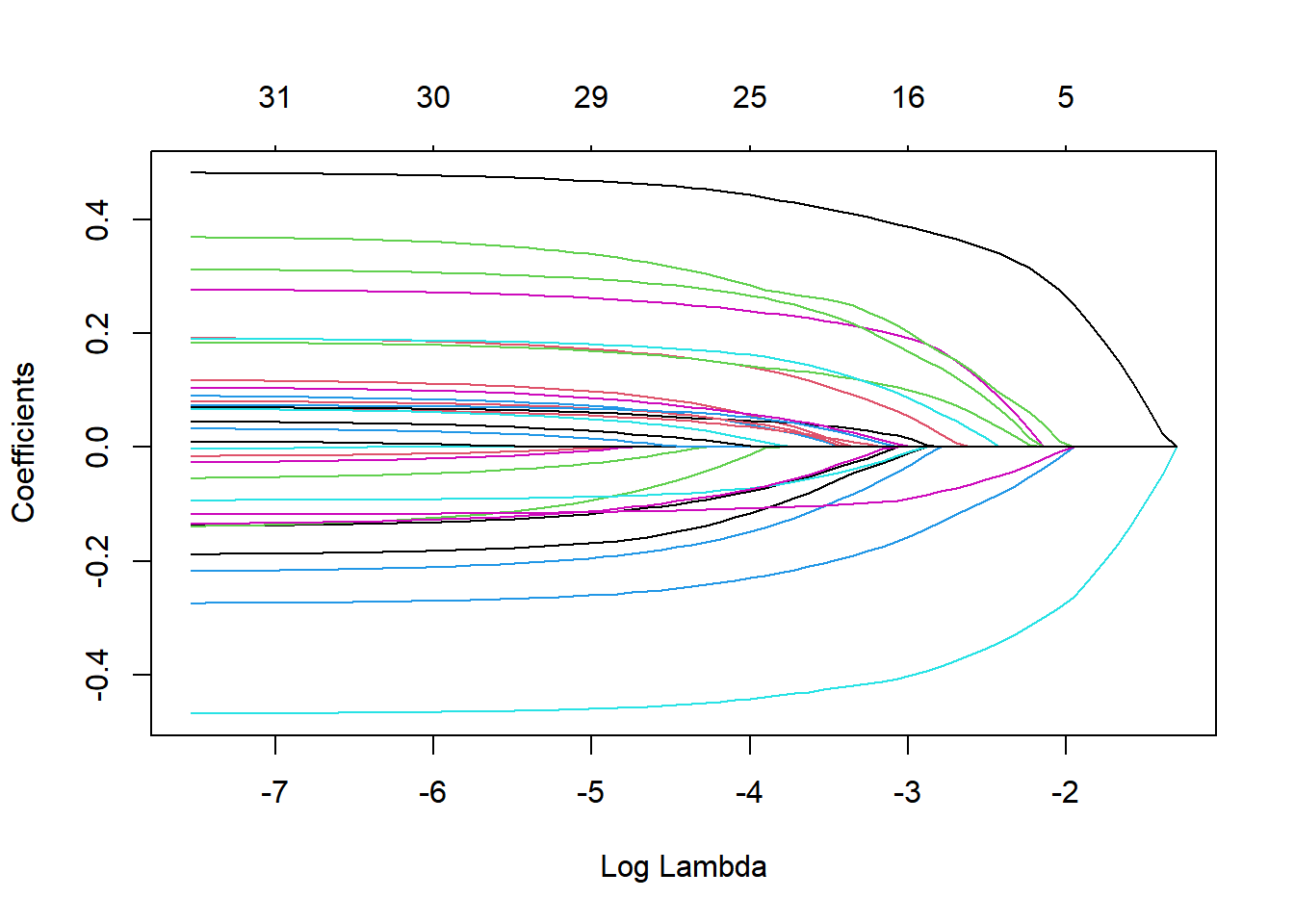
lasso_best_fit%>%
extract_fit_parsnip()%>%
tidy()%>%
filter(estimate!="0") #several estimates are at 0, ignore.## # A tibble: 13 x 3
## term estimate penalty
## <chr> <dbl> <dbl>
## 1 (Intercept) 98.7 0.0574
## 2 ChestCongestion_Yes 0.0332 0.0574
## 3 ChillsSweats_Yes 0.0894 0.0574
## 4 NasalCongestion_Yes -0.140 0.0574
## 5 Sneeze_Yes -0.391 0.0574
## 6 Fatigue_Yes 0.178 0.0574
## 7 SubjectiveFever_Yes 0.377 0.0574
## 8 Weakness_1 0.178 0.0574
## 9 Myalgia_2 -0.00994 0.0574
## 10 Myalgia_3 0.0679 0.0574
## 11 RunnyNose_Yes -0.0825 0.0574
## 12 Nausea_Yes 0.00349 0.0574
## 13 Pharyngitis_Yes 0.148 0.0574#predicted versus observed from tuned model
plot(lasso_pred$.pred,train_data$BodyTemp,
xlim =c(97,104),
ylim=c(97,104))
abline(a=0,b=1, col = 'red') #45 degree line, along which the results should fall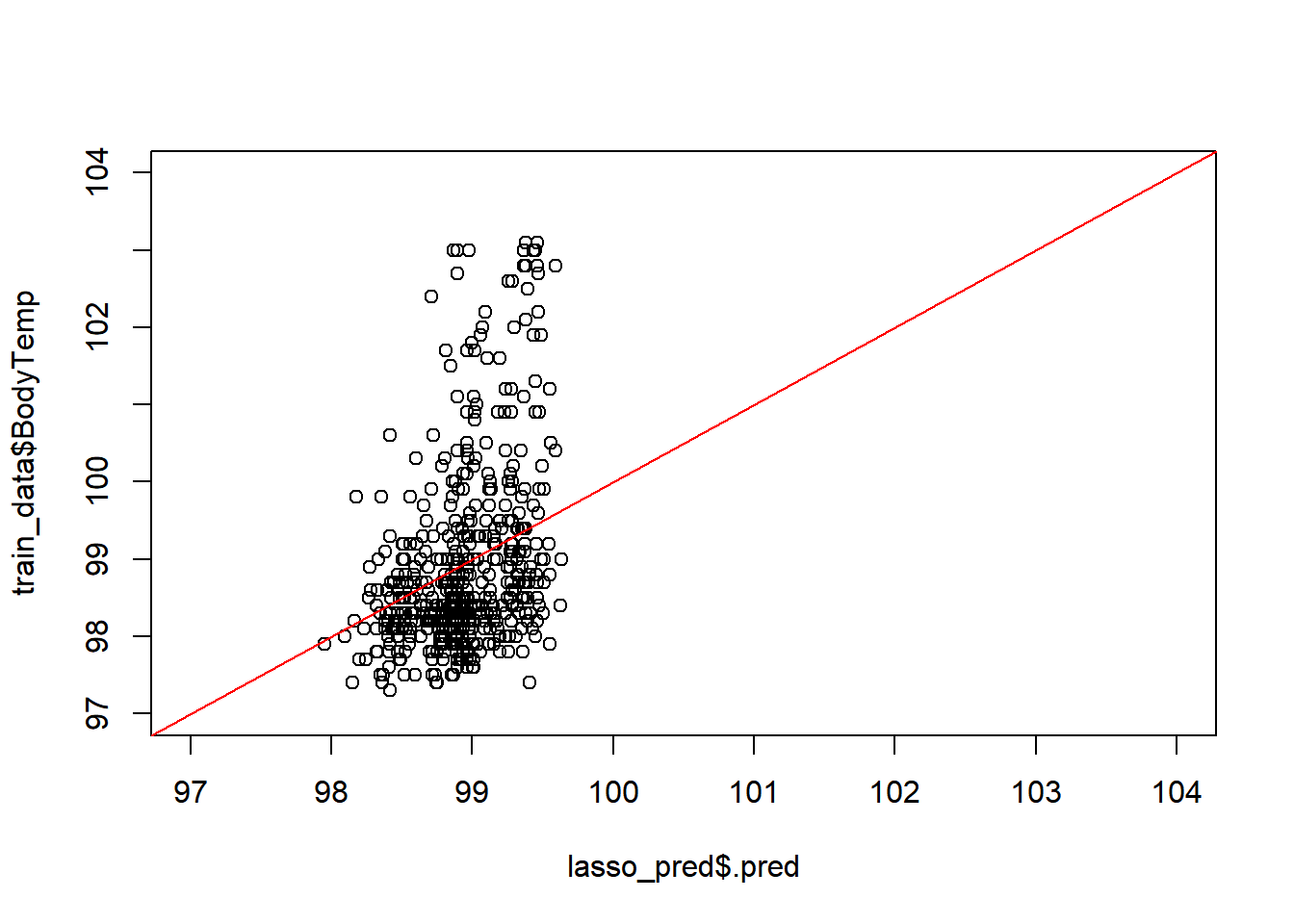
#residuals
plot(lasso_pred$.pred-train_data$BodyTemp)
abline(a=0,b=0, col = 'red') 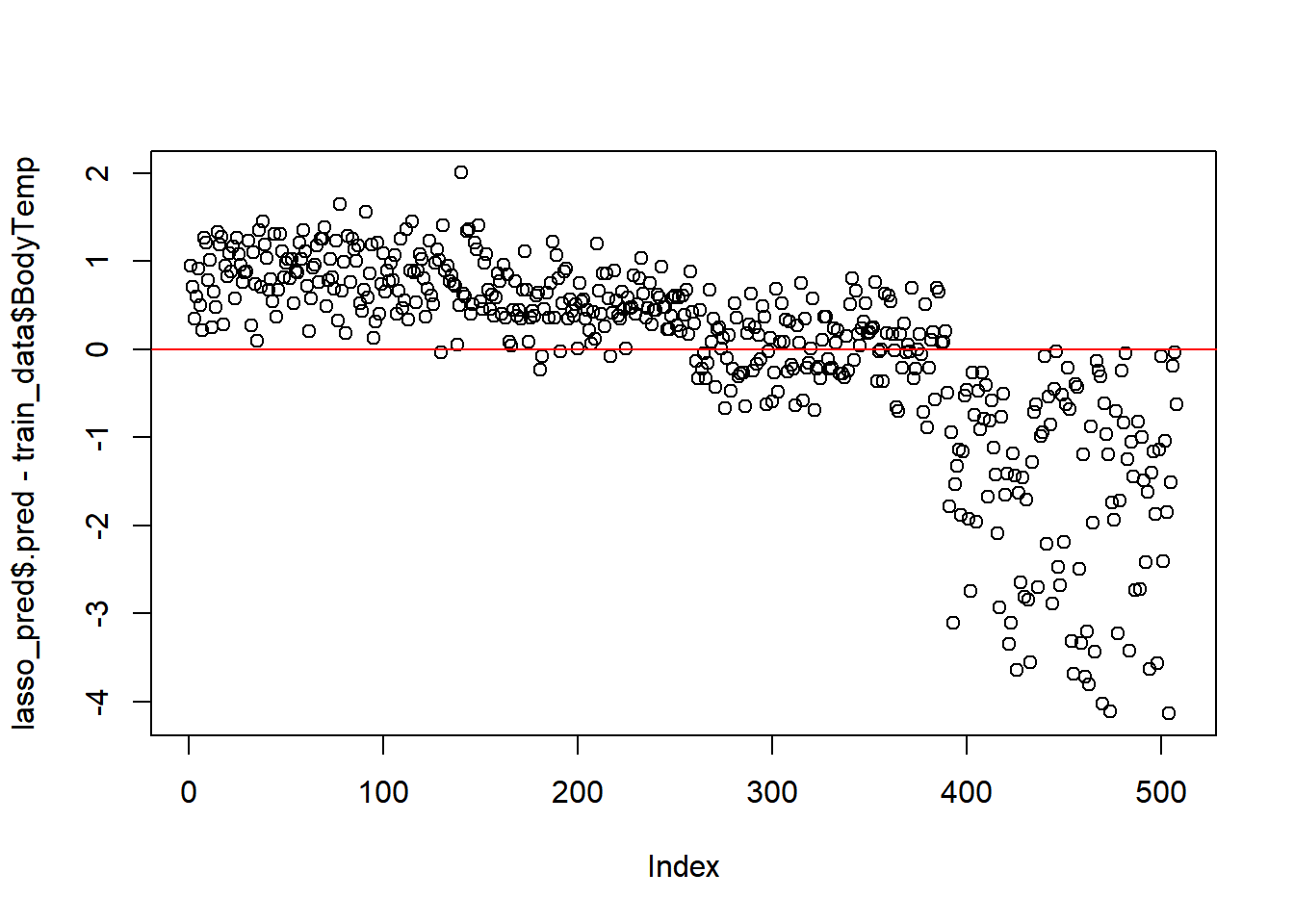
Ideally, good results should fall along the red line.
#LASSO Model Performance
lasso_perfomance <-
lasso_tune_rec %>%
show_best(n = 1)%>%
print()## # A tibble: 1 x 7
## penalty .metric .estimator mean n std_err .config
## <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 0.0574 rmse standard 1.15 25 0.0169 Preprocessor1_Model18#View RMSE comparison with nulls
lasso_RMSE<-
lasso_tune_rec%>%
show_best(n=1)%>%
transmute(
rmse= round(mean,2),
SE= round(std_err,2),
model="LASSO"
)LASSO RMSE mean is lower, just a little, from null train and test values.
Random Forest (RF)
NOTE: Aspects from the Decision Tree Model and LASSO Linear Regressionwill be used to create a Random Forest Model. Code for this section may be taken from TidyModels Tutorial Case Study.
RF Setup
RF_model <-
rand_forest() %>%
set_args(mtry = tune(),
trees = tune(),
min_n = tune()
) %>%
# select the engine/package that underlies the model
set_engine("ranger",
num.threads = 18,
#for some reason for RF, we need to set this in the engine too
importance = "permutation") %>%
# choose either the continuous regression or binary classification mode
set_mode("regression")
#view identified parameters to be tuned
RF_model%>%
parameters()## Collection of 3 parameters for tuning
##
## identifier type object
## mtry mtry nparam[?]
## trees trees nparam[+]
## min_n min_n nparam[+]
##
## Model parameters needing finalization:
## # Randomly Selected Predictors ('mtry')
##
## See `?dials::finalize` or `?dials::update.parameters` for more information.RF WorkFlow
# regression workflow
RF_WF <-
workflow() %>%
add_model(RF_model) %>%
add_recipe(Mod11_rec)
print(RF_WF)## == Workflow ====================================================================
## Preprocessor: Recipe
## Model: rand_forest()
##
## -- Preprocessor ----------------------------------------------------------------
## 1 Recipe Step
##
## * step_dummy()
##
## -- Model -----------------------------------------------------------------------
## Random Forest Model Specification (regression)
##
## Main Arguments:
## mtry = tune()
## trees = tune()
## min_n = tune()
##
## Engine-Specific Arguments:
## num.threads = 18
## importance = permutation
##
## Computational engine: rangerRF Tuning
#tuning grid
RF_grid<-
expand.grid(mtry = c(3, 4, 5, 6),
min_n = c(40, 50, 60),
trees = c(500,1000))RF_tune_rec<-
RF_WF%>%
tune_grid(resamples = folds, #tune with CV value pre designated
grid = RF_grid, #just created grid of values
control = control_grid(
save_pred = TRUE
),
metrics = metric_set(rmse) #RMSE as target metric
)RF Evaluation
Autoplot
RF_tune_rec%>%
autoplot()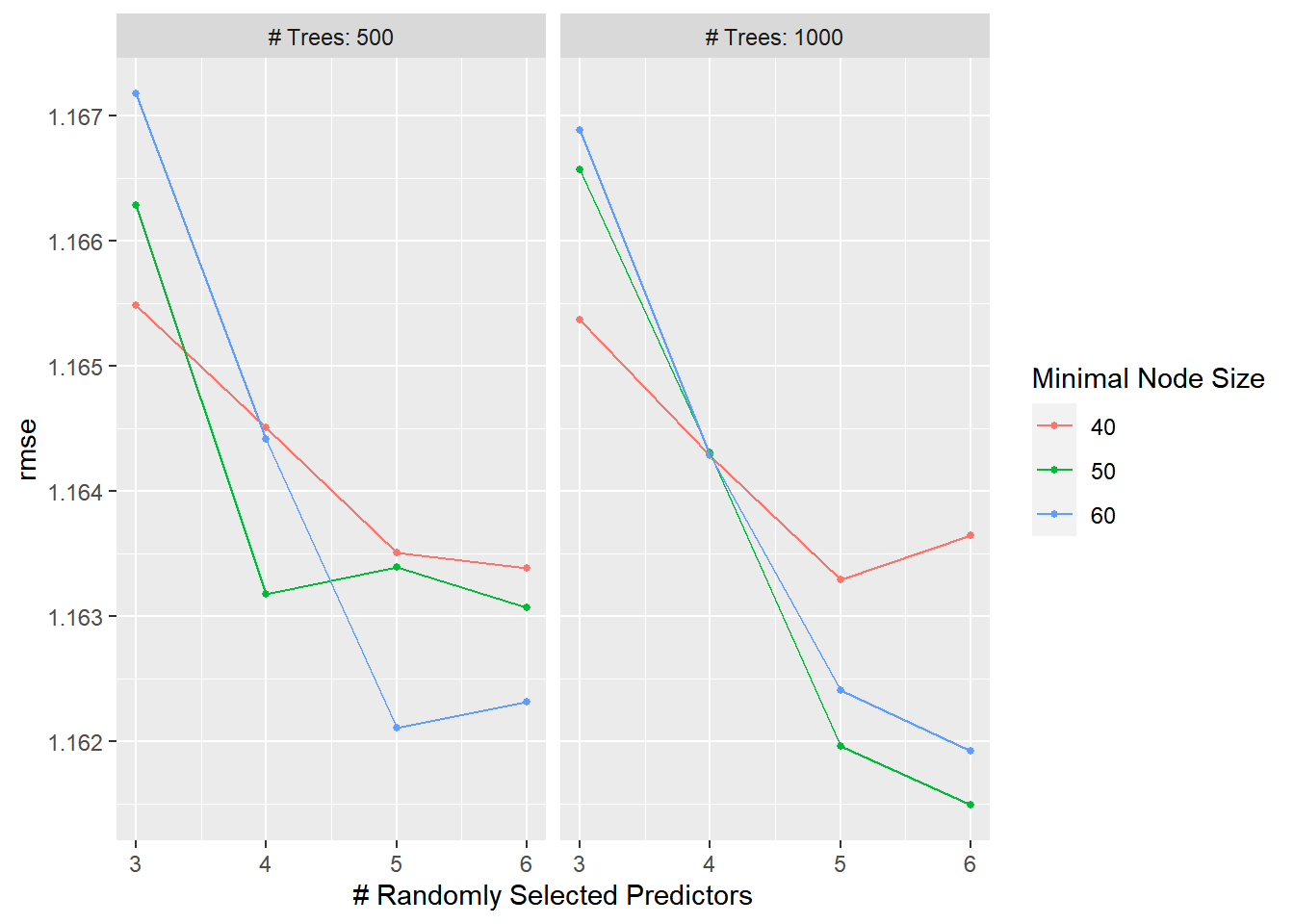
# get the tuned model that performs best
RF_best <-
RF_tune_rec %>%
select_best(metric = "rmse")
# finalize workflow with best model
RF_best_WF <-
RF_WF %>%
finalize_workflow(RF_best)
# fitting best performing model
RF_best_fit <-
RF_best_WF %>%
fit(data = train_data)
RF_pred <-
predict(RF_best_fit,
train_data)Unfortunately, there’s not an easy way to look at a random forest model. Below are examples at looking at the data.
View importance predictors by using vip.
#pull out the fit object
RF_x <- RF_best_fit$fit$fit$fit
#plot variable importance
vip(RF_x, num_features = 20) #can specify features, default is 10.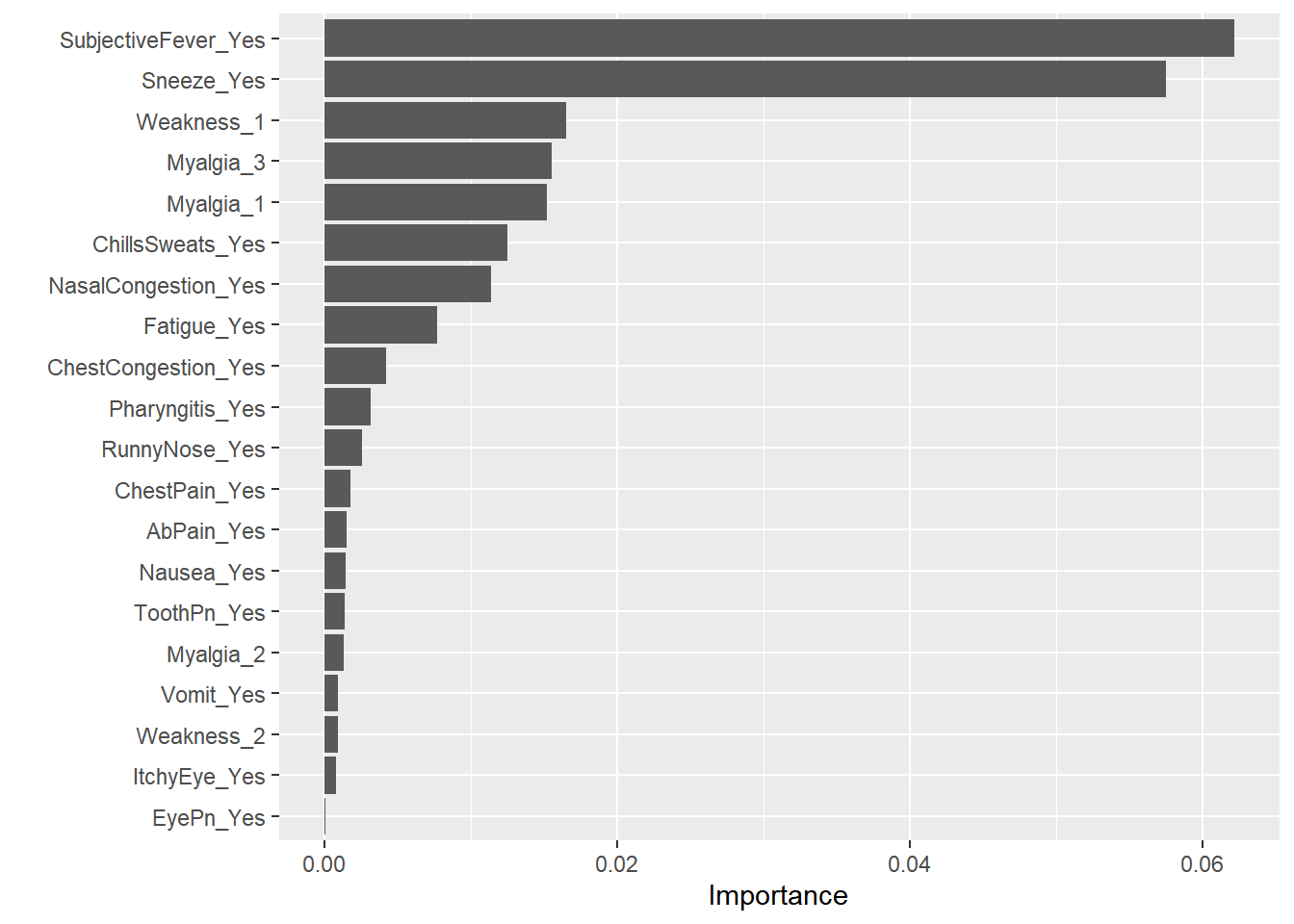 This makes perfect sense that a fever would indicate a difference in BodyTemp– Body Temperature
This makes perfect sense that a fever would indicate a difference in BodyTemp– Body Temperature
Plots:
Predicted vs Observed
plot(RF_pred$.pred,
train_data$BodyTemp,
xlim =c(97,103), #x= Actual Body Temp,
ylim=c(97,103)) # y= predicted body temp
abline(a=0,b=1, col = 'red') 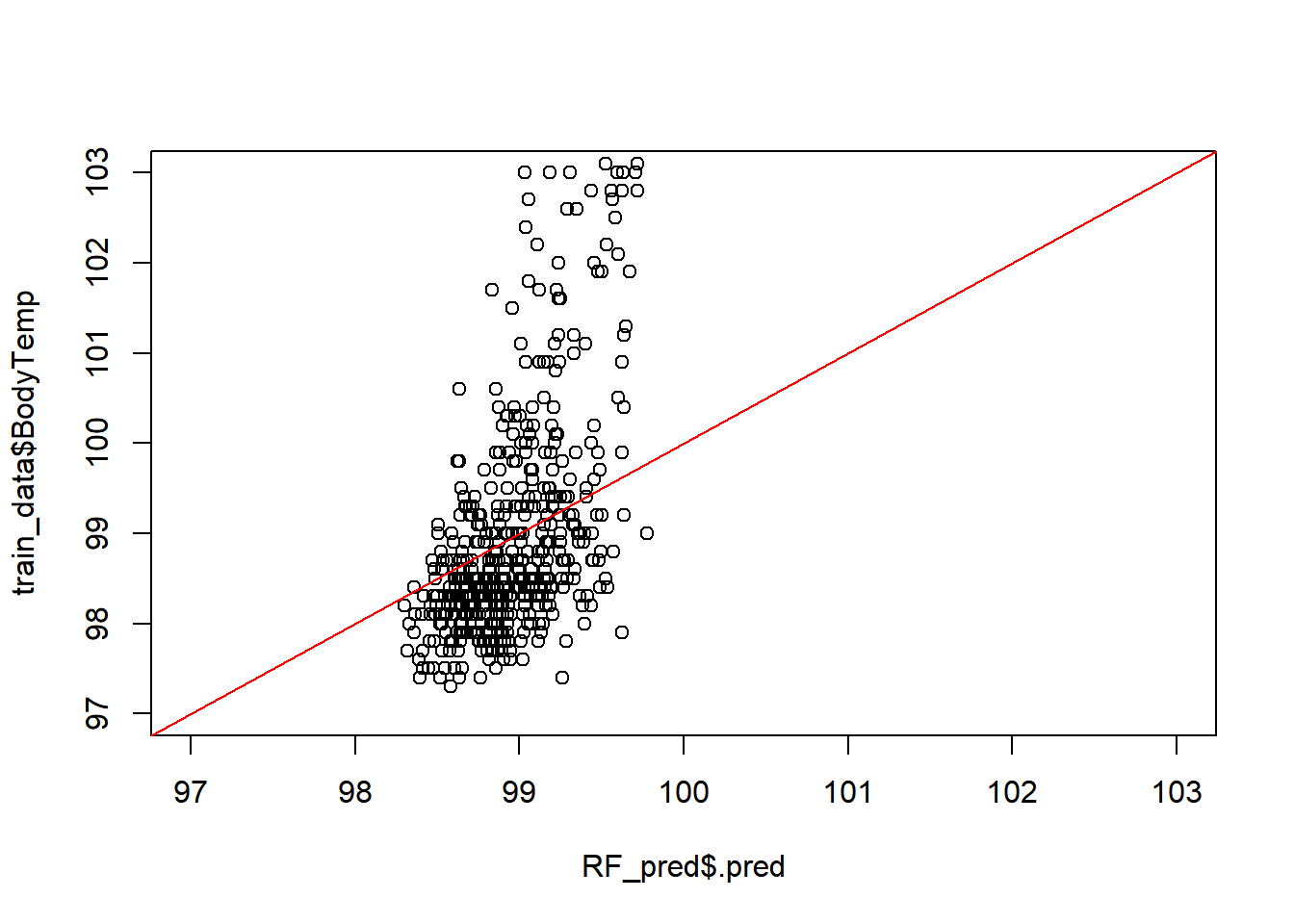
#45 degree line, along which the results should fallModel fit vs Residuals
#residuals
plot(RF_pred$.pred-train_data$BodyTemp) # Index= Observation Number
#straight line, along which the results should fall
abline(a=0,b=0, col = 'red') 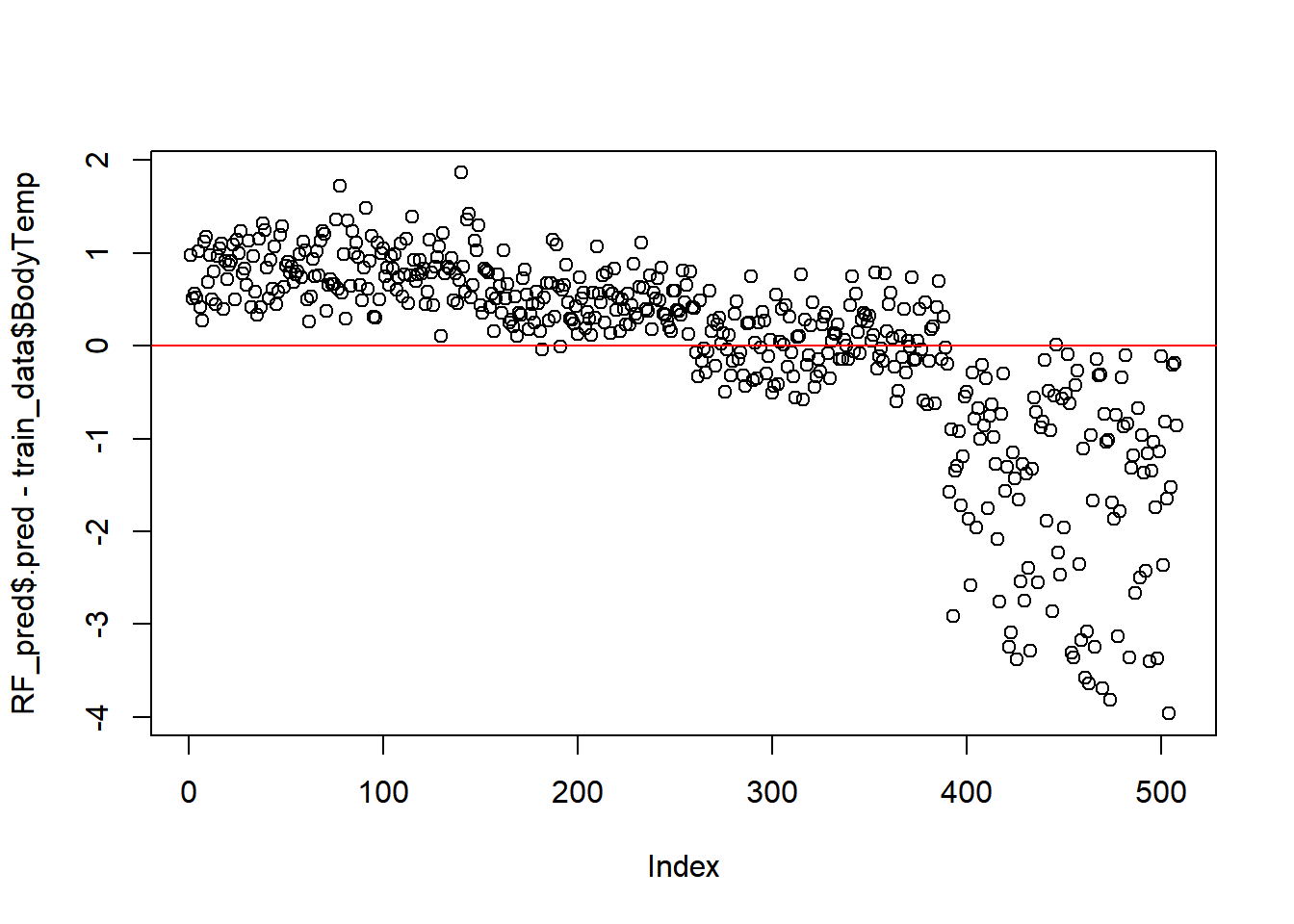
Model Performance
RF_perfomance <-
RF_tune_rec %>%
show_best(n = 1)
print(RF_perfomance) ## # A tibble: 1 x 9
## mtry trees min_n .metric .estimator mean n std_err .config
## <dbl> <dbl> <dbl> <chr> <chr> <dbl> <int> <dbl> <chr>
## 1 6 1000 50 rmse standard 1.16 25 0.0168 Preprocessor1_Model20#View RMSE comparison with nulls
RF_RMSE<-
RF_tune_rec%>%
show_best(n=1)%>%
transmute(
rmse= round(mean,2),
SE= round(std_err,2),
model="RF"
)Compared with the nulls this is really no better than the null rmse
Final Model Selection and Evaluation
Compare all RMSE(s) from the models to the Nulls
CompareRMSE<-
bind_rows(tree_RMSE)%>%
bind_rows(lasso_RMSE)%>%
bind_rows(RF_RMSE)%>%
bind_rows(null_RMSE_train)%>%
mutate(rmse=round(rmse,2))%>%
arrange(rmse)%>% #arrange RMSE that is the smallest
print()## # A tibble: 4 x 3
## rmse SE model
## <dbl> <dbl> <chr>
## 1 1.15 0.02 LASSO
## 2 1.16 0.02 RF
## 3 1.19 0.02 Tree
## 4 1.21 0 Null - TrainNone of the models fit the data well. This suggests that the predictor variables in the data set may not be useful in predicting the body temperature of a suspect flu case (outcome).
LASSO model is the best model based on RMSE value being the lowest.
As such, the Lasso model will be evaluated by fitting the LASSO model to the training_data set and evaluate on the test_data.
Performance Check
Fit_Test<-
lasso_best_WF%>% # wf of the best fit from the training data
last_fit(split=data_split)Compare test against training
final_fit <-
lasso_best_WF%>%
last_fit(data_split)
Final_performance<-
final_fit%>%
collect_metrics()%>%
print()## # A tibble: 2 x 4
## .metric .estimator .estimate .config
## <chr> <chr> <dbl> <chr>
## 1 rmse standard 1.15 Preprocessor1_Model1
## 2 rsq standard 0.0291 Preprocessor1_Model1test_predictions <-
final_fit %>%
collect_predictions()Predicted vs Observed
plot(test_predictions$.pred,
test_data$BodyTemp, #changeed to test data, previously used training
xlim =c(97,103), #x= Actual Body Temp,
ylim=c(97,103)) # y= predicted body temp
abline(a=0,b=1, col = 'red') 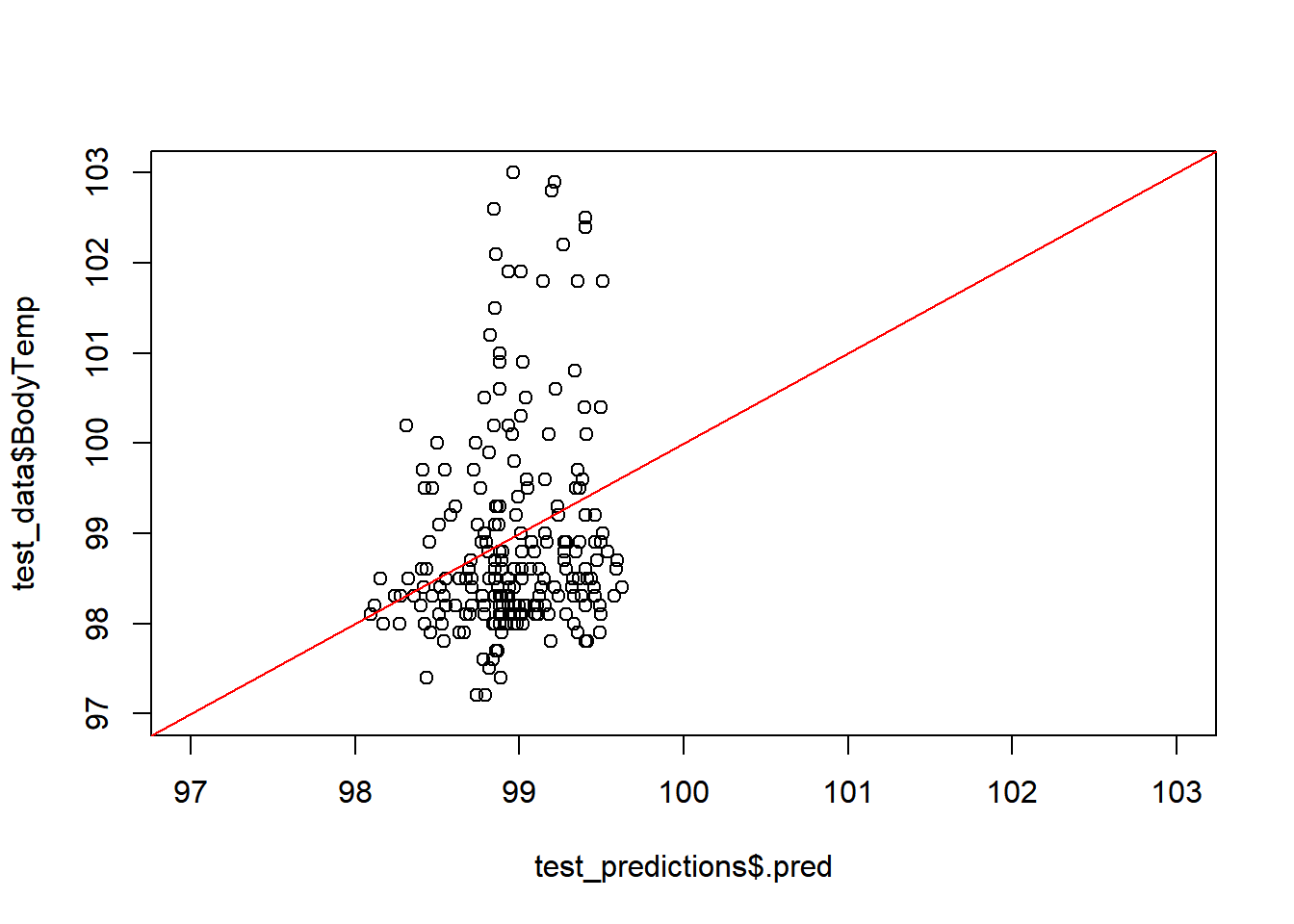
#45 degree line, along which the results should fallModel fit vs Residuals
#residuals
plot(test_predictions$.pred-test_data$BodyTemp) # Index= Observation Number
#straight line, along which the results should fall
abline(a=0,b=0, col = 'red') 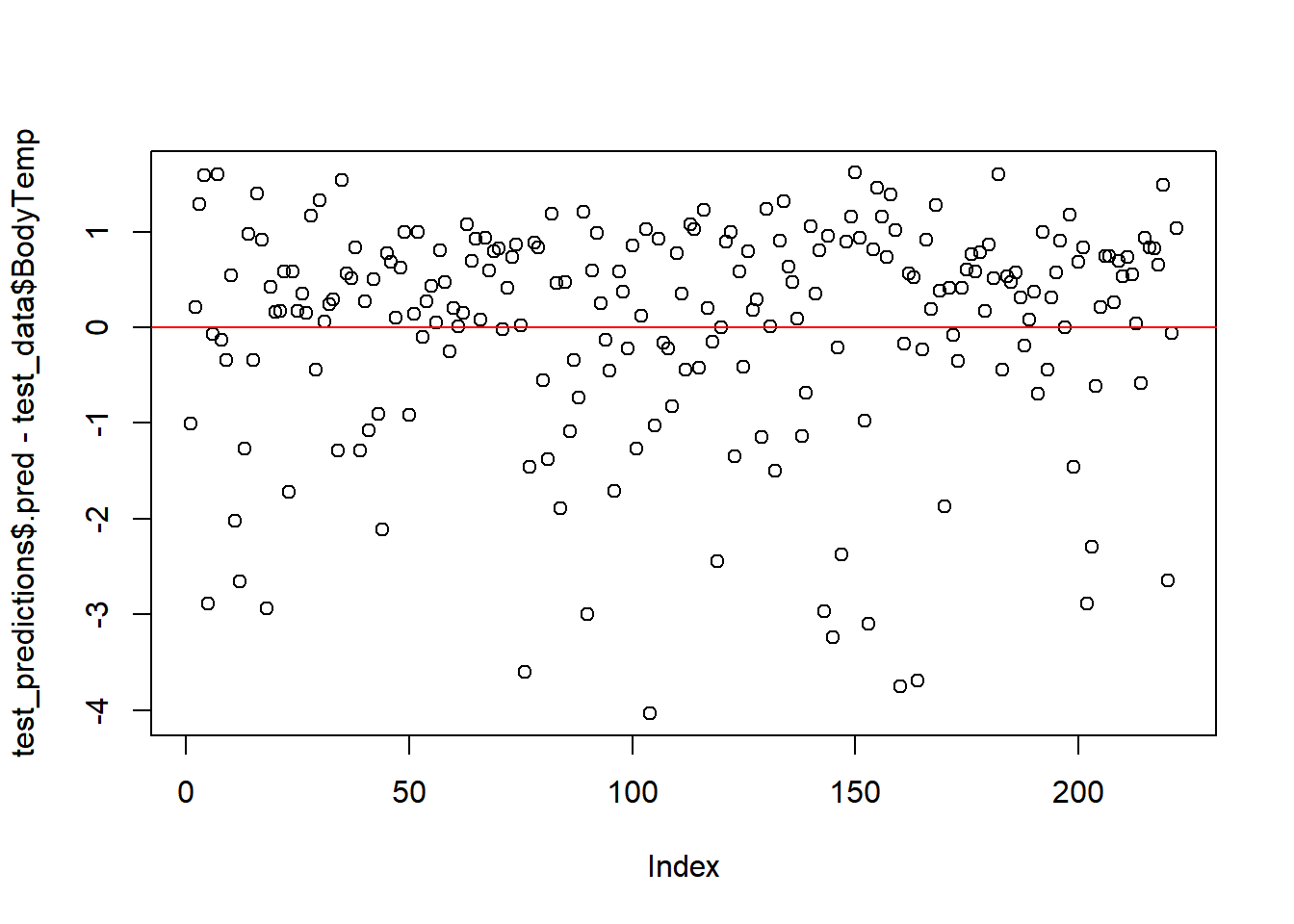 # Comments
# Comments
LASSO was the best selection from the model types used in this assessment. But it still isn’t great. This suggests that the predictors are not great at predicting the outcome. From the importance predictors the most related to body temp is fever, which makes sense, but in terms of predicting flu– it still has some uncertainty. As there are people who may be positive for flu but may not have severe enough symptoms to warrant getting treated and thus by passing the collection of symptoms.