R coding Exercise
This will contain the future R coding exercise.
Loading and checking data data from dslabs package and gapminder
library(dslabs)
#library(gapminder)
library(tidyverse)
#summary(data(gapminder))
### above is what caused my weird data
#look at help files for gapminder
#help(gapminder)
#overview of data structure
str(gapminder)## 'data.frame': 10545 obs. of 9 variables:
## $ country : Factor w/ 185 levels "Albania","Algeria",..: 1 2 3 4 5 6 7 8 9 10 ...
## $ year : int 1960 1960 1960 1960 1960 1960 1960 1960 1960 1960 ...
## $ infant_mortality: num 115.4 148.2 208 NA 59.9 ...
## $ life_expectancy : num 62.9 47.5 36 63 65.4 ...
## $ fertility : num 6.19 7.65 7.32 4.43 3.11 4.55 4.82 3.45 2.7 5.57 ...
## $ population : num 1636054 11124892 5270844 54681 20619075 ...
## $ gdp : num NA 1.38e+10 NA NA 1.08e+11 ...
## $ continent : Factor w/ 5 levels "Africa","Americas",..: 4 1 1 2 2 3 2 5 4 3 ...
## $ region : Factor w/ 22 levels "Australia and New Zealand",..: 19 11 10 2 15 21 2 1 22 21 ...#summary of data
summary(gapminder)## country year infant_mortality life_expectancy
## Albania : 57 Min. :1960 Min. : 1.50 Min. :13.20
## Algeria : 57 1st Qu.:1974 1st Qu.: 16.00 1st Qu.:57.50
## Angola : 57 Median :1988 Median : 41.50 Median :67.54
## Antigua and Barbuda: 57 Mean :1988 Mean : 55.31 Mean :64.81
## Argentina : 57 3rd Qu.:2002 3rd Qu.: 85.10 3rd Qu.:73.00
## Armenia : 57 Max. :2016 Max. :276.90 Max. :83.90
## (Other) :10203 NA's :1453
## fertility population gdp continent
## Min. :0.840 Min. :3.124e+04 Min. :4.040e+07 Africa :2907
## 1st Qu.:2.200 1st Qu.:1.333e+06 1st Qu.:1.846e+09 Americas:2052
## Median :3.750 Median :5.009e+06 Median :7.794e+09 Asia :2679
## Mean :4.084 Mean :2.701e+07 Mean :1.480e+11 Europe :2223
## 3rd Qu.:6.000 3rd Qu.:1.523e+07 3rd Qu.:5.540e+10 Oceania : 684
## Max. :9.220 Max. :1.376e+09 Max. :1.174e+13
## NA's :187 NA's :185 NA's :2972
## region
## Western Asia :1026
## Eastern Africa : 912
## Western Africa : 912
## Caribbean : 741
## South America : 684
## Southern Europe: 684
## (Other) :5586#determine type of object gapminder is
class(gapminder)## [1] "data.frame"Processing Data w/tidyverse
library(tidyverse)
library(dplyr)#making africadata from gapminder
africadata<-gapminder%>%
filter(continent=="Africa")#checking africadata
str(africadata)## 'data.frame': 2907 obs. of 9 variables:
## $ country : Factor w/ 185 levels "Albania","Algeria",..: 2 3 18 22 26 27 29 31 32 33 ...
## $ year : int 1960 1960 1960 1960 1960 1960 1960 1960 1960 1960 ...
## $ infant_mortality: num 148 208 187 116 161 ...
## $ life_expectancy : num 47.5 36 38.3 50.3 35.2 ...
## $ fertility : num 7.65 7.32 6.28 6.62 6.29 6.95 5.65 6.89 5.84 6.25 ...
## $ population : num 11124892 5270844 2431620 524029 4829291 ...
## $ gdp : num 1.38e+10 NA 6.22e+08 1.24e+08 5.97e+08 ...
## $ continent : Factor w/ 5 levels "Africa","Americas",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ region : Factor w/ 22 levels "Australia and New Zealand",..: 11 10 20 17 20 5 10 20 10 10 ...summary(africadata)## country year infant_mortality life_expectancy
## Algeria : 57 Min. :1960 Min. : 11.40 Min. :13.20
## Angola : 57 1st Qu.:1974 1st Qu.: 62.20 1st Qu.:48.23
## Benin : 57 Median :1988 Median : 93.40 Median :53.98
## Botswana : 57 Mean :1988 Mean : 95.12 Mean :54.38
## Burkina Faso: 57 3rd Qu.:2002 3rd Qu.:124.70 3rd Qu.:60.10
## Burundi : 57 Max. :2016 Max. :237.40 Max. :77.60
## (Other) :2565 NA's :226
## fertility population gdp continent
## Min. :1.500 Min. : 41538 Min. :4.659e+07 Africa :2907
## 1st Qu.:5.160 1st Qu.: 1605232 1st Qu.:8.373e+08 Americas: 0
## Median :6.160 Median : 5570982 Median :2.448e+09 Asia : 0
## Mean :5.851 Mean : 12235961 Mean :9.346e+09 Europe : 0
## 3rd Qu.:6.860 3rd Qu.: 13888152 3rd Qu.:6.552e+09 Oceania : 0
## Max. :8.450 Max. :182201962 Max. :1.935e+11
## NA's :51 NA's :51 NA's :637
## region
## Eastern Africa :912
## Western Africa :912
## Middle Africa :456
## Northern Africa :342
## Southern Africa :285
## Australia and New Zealand: 0
## (Other) : 0class(africadata)## [1] "data.frame"#separate africadata into 2 new variables
Ainfant_mortality<-africadata%>%
select(infant_mortality)
str(Ainfant_mortality)## 'data.frame': 2907 obs. of 1 variable:
## $ infant_mortality: num 148 208 187 116 161 ...summary(Ainfant_mortality)## infant_mortality
## Min. : 11.40
## 1st Qu.: 62.20
## Median : 93.40
## Mean : 95.12
## 3rd Qu.:124.70
## Max. :237.40
## NA's :226Alife_expectancy<-africadata%>%
select(life_expectancy)
str(Alife_expectancy)## 'data.frame': 2907 obs. of 1 variable:
## $ life_expectancy: num 47.5 36 38.3 50.3 35.2 ...summary(Alife_expectancy)## life_expectancy
## Min. :13.20
## 1st Qu.:48.23
## Median :53.98
## Mean :54.38
## 3rd Qu.:60.10
## Max. :77.60Plotting
#Plotting infant_mortality
africadata%>%
ggplot(aes(infant_mortality, population))+
geom_point(aes(color=year, shape=region))## Warning: Removed 226 rows containing missing values (geom_point).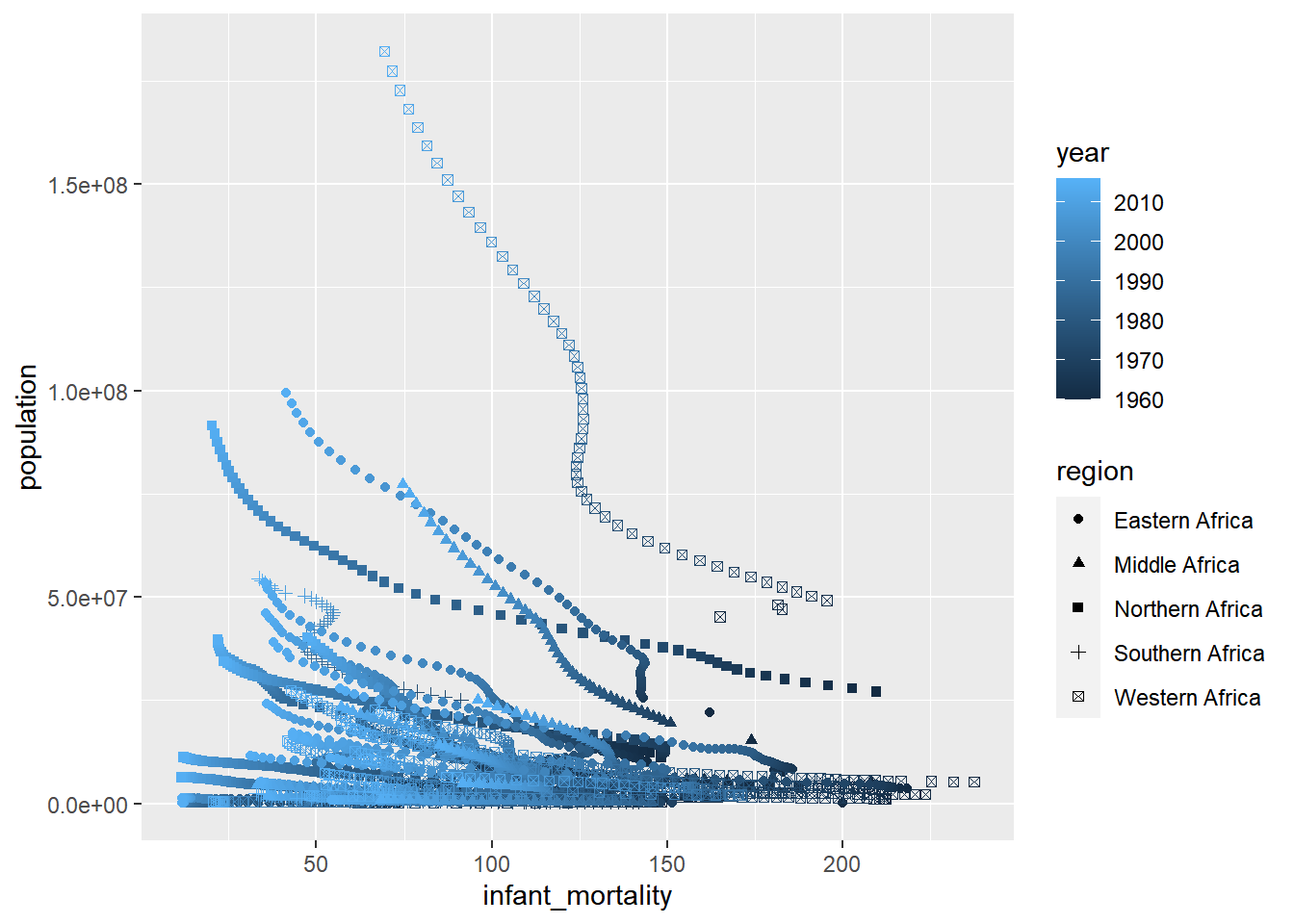 #Plotting life expectancy
#Plotting life expectancy
africadata%>%
ggplot(aes(life_expectancy, population))+
geom_point(aes(shape=region, color=year))## Warning: Removed 51 rows containing missing values (geom_point).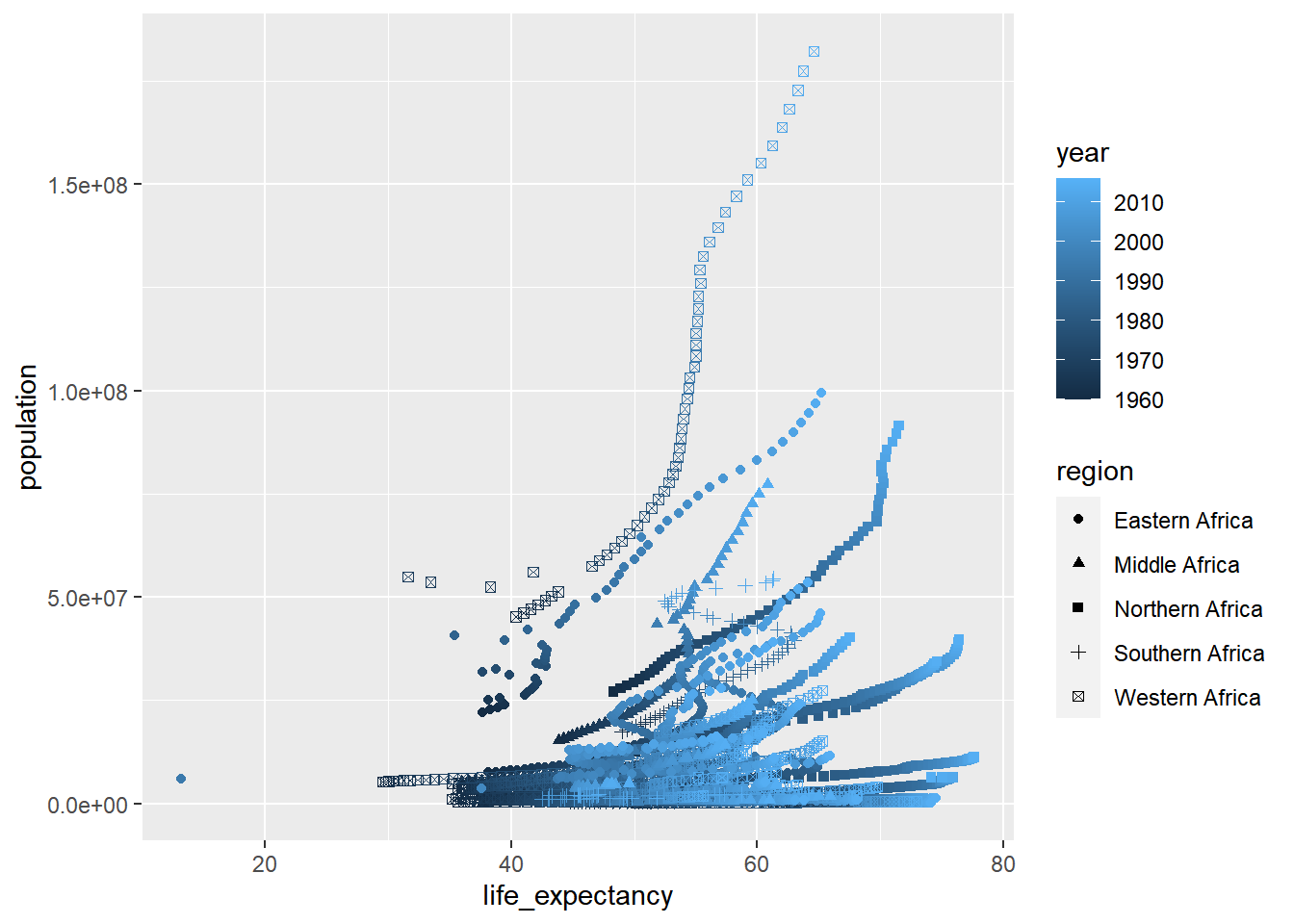 Both plots appear to have merging dots scattered across the population and expectancy. This is likely due to not parsing out the countries within Africa, the year the data was taken from, or region… depends on what we want to look for.
Both plots appear to have merging dots scattered across the population and expectancy. This is likely due to not parsing out the countries within Africa, the year the data was taken from, or region… depends on what we want to look for.
More data processing
summary(africadata$year)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 1960 1974 1988 1988 2002 2016#filter missing values (NA) for infant mortality
africadata%>%
select(year, infant_mortality)%>%
filter(is.na(infant_mortality))%>%
count(year)## year n
## 1 1960 10
## 2 1961 17
## 3 1962 16
## 4 1963 16
## 5 1964 15
## 6 1965 14
## 7 1966 13
## 8 1967 11
## 9 1968 11
## 10 1969 7
## 11 1970 5
## 12 1971 6
## 13 1972 6
## 14 1973 6
## 15 1974 5
## 16 1975 5
## 17 1976 3
## 18 1977 3
## 19 1978 2
## 20 1979 2
## 21 1980 1
## 22 1981 1
## 23 2016 51#filter for year 2000
AD2K<-africadata%>%
filter(year=="2000")
summary(AD2K)## country year infant_mortality life_expectancy
## Algeria : 1 Min. :2000 Min. : 12.30 Min. :37.60
## Angola : 1 1st Qu.:2000 1st Qu.: 60.80 1st Qu.:51.75
## Benin : 1 Median :2000 Median : 80.30 Median :54.30
## Botswana : 1 Mean :2000 Mean : 78.93 Mean :56.36
## Burkina Faso: 1 3rd Qu.:2000 3rd Qu.:103.30 3rd Qu.:60.00
## Burundi : 1 Max. :2000 Max. :143.30 Max. :75.00
## (Other) :45
## fertility population gdp continent
## Min. :1.990 Min. : 81154 Min. :2.019e+08 Africa :51
## 1st Qu.:4.150 1st Qu.: 2304687 1st Qu.:1.274e+09 Americas: 0
## Median :5.550 Median : 8799165 Median :3.238e+09 Asia : 0
## Mean :5.156 Mean : 15659800 Mean :1.155e+10 Europe : 0
## 3rd Qu.:5.960 3rd Qu.: 17391242 3rd Qu.:8.654e+09 Oceania : 0
## Max. :7.730 Max. :122876723 Max. :1.329e+11
##
## region
## Eastern Africa :16
## Western Africa :16
## Middle Africa : 8
## Northern Africa : 6
## Southern Africa : 5
## Australia and New Zealand: 0
## (Other) : 0str(AD2K)## 'data.frame': 51 obs. of 9 variables:
## $ country : Factor w/ 185 levels "Albania","Algeria",..: 2 3 18 22 26 27 29 31 32 33 ...
## $ year : int 2000 2000 2000 2000 2000 2000 2000 2000 2000 2000 ...
## $ infant_mortality: num 33.9 128.3 89.3 52.4 96.2 ...
## $ life_expectancy : num 73.3 52.3 57.2 47.6 52.6 46.7 54.3 68.4 45.3 51.5 ...
## $ fertility : num 2.51 6.84 5.98 3.41 6.59 7.06 5.62 3.7 5.45 7.35 ...
## $ population : num 31183658 15058638 6949366 1736579 11607944 ...
## $ gdp : num 5.48e+10 9.13e+09 2.25e+09 5.63e+09 2.61e+09 ...
## $ continent : Factor w/ 5 levels "Africa","Americas",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ region : Factor w/ 22 levels "Australia and New Zealand",..: 11 10 20 17 20 5 10 20 10 10 ...More Plotting
#Fit1 - life expectancy as outcome and infant mortality as predictor
Fit1<- lm(AD2K$life_expectancy~AD2K$infant_mortality)
summary(Fit1)##
## Call:
## lm(formula = AD2K$life_expectancy ~ AD2K$infant_mortality)
##
## Residuals:
## Min 1Q Median 3Q Max
## -22.6651 -3.7087 0.9914 4.0408 8.6817
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 71.29331 2.42611 29.386 < 2e-16 ***
## AD2K$infant_mortality -0.18916 0.02869 -6.594 2.83e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 6.221 on 49 degrees of freedom
## Multiple R-squared: 0.4701, Adjusted R-squared: 0.4593
## F-statistic: 43.48 on 1 and 49 DF, p-value: 2.826e-08#Fit2 life expectancy as outcome, pop size as predictor
Fit2<- lm(AD2K$life_expectancy~AD2K$population)
summary(Fit2)##
## Call:
## lm(formula = AD2K$life_expectancy ~ AD2K$population)
##
## Residuals:
## Min 1Q Median 3Q Max
## -18.429 -4.602 -2.568 3.800 18.802
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 5.593e+01 1.468e+00 38.097 <2e-16 ***
## AD2K$population 2.756e-08 5.459e-08 0.505 0.616
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 8.524 on 49 degrees of freedom
## Multiple R-squared: 0.005176, Adjusted R-squared: -0.01513
## F-statistic: 0.2549 on 1 and 49 DF, p-value: 0.6159Thoughts on the p-values. Fit 1 is significantly different so there are effects between life expectancy and infant mortality in 2000. Fit 2 does not show a significant difference so no real effects on the life expectancy with the population in 2000.
#Additional analysis by Priyanka Gannavarapu
Relationship between region and life expectancy
ggplot(AD2K, aes(x= region, y = life_expectancy)) +
geom_boxplot() 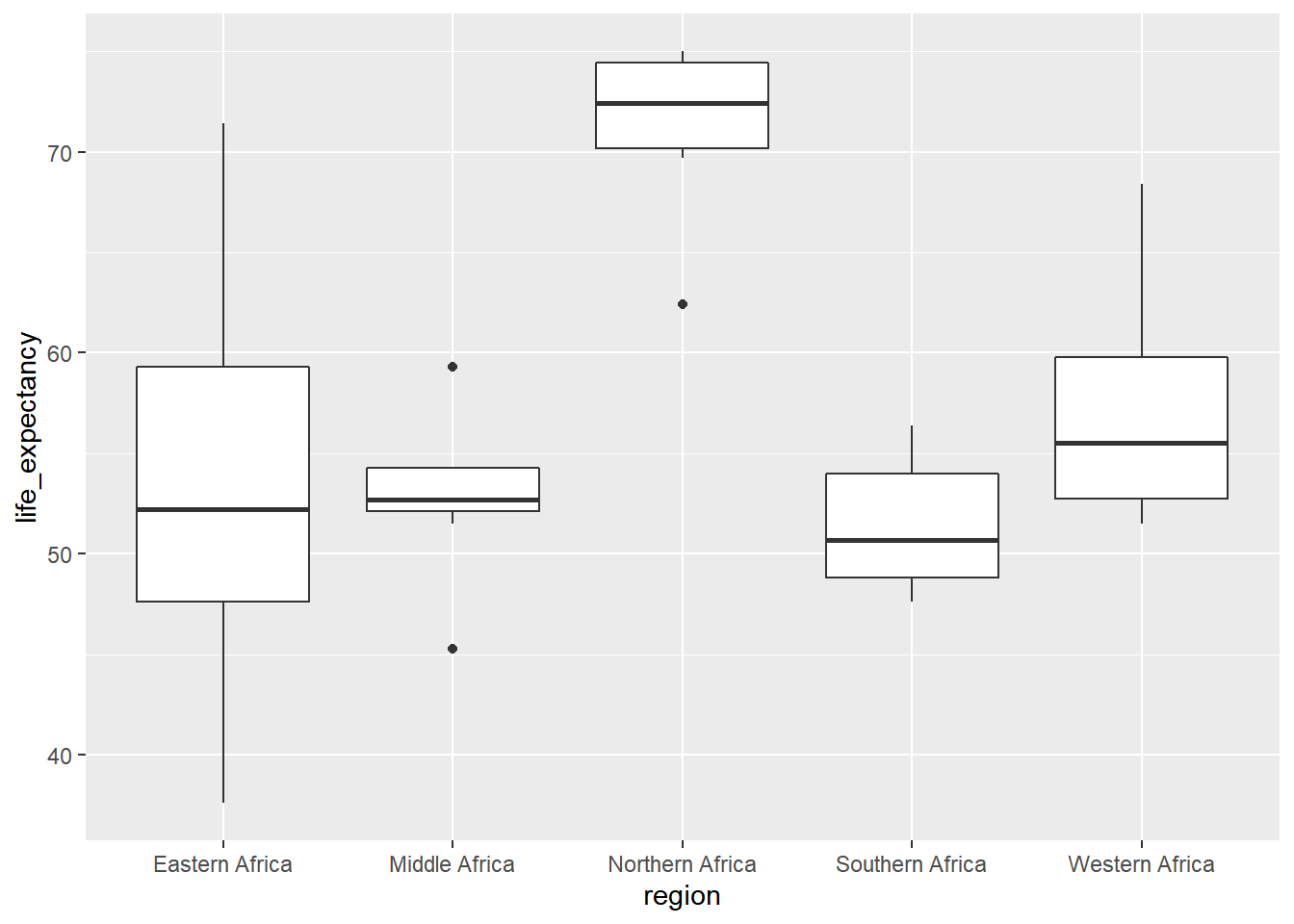
Fitting two Linear models .
fit3 = lm(life_expectancy ~ gdp, data= AD2K)
summary(fit3)##
## Call:
## lm(formula = life_expectancy ~ gdp, data = AD2K)
##
## Residuals:
## Min 1Q Median 3Q Max
## -17.488 -4.316 -1.890 4.272 17.479
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 5.501e+01 1.247e+00 44.106 <2e-16 ***
## gdp 1.168e-10 4.640e-11 2.516 0.0152 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 8.043 on 49 degrees of freedom
## Multiple R-squared: 0.1144, Adjusted R-squared: 0.09632
## F-statistic: 6.329 on 1 and 49 DF, p-value: 0.0152#The model is statistically significant.
fit4 = lm(life_expectancy~ infant_mortality + region , data = AD2K)
summary(fit4)##
## Call:
## lm(formula = life_expectancy ~ infant_mortality + region, data = AD2K)
##
## Residuals:
## Min 1Q Median 3Q Max
## -19.1675 -2.4682 0.6053 2.2362 7.8495
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 66.95785 2.36426 28.321 < 2e-16 ***
## infant_mortality -0.17479 0.02694 -6.488 5.87e-08 ***
## regionMiddle Africa 2.96949 2.12310 1.399 0.16877
## regionNorthern Africa 10.89992 2.47117 4.411 6.35e-05 ***
## regionSouthern Africa -4.13489 2.43244 -1.700 0.09605 .
## regionWestern Africa 5.92329 1.71809 3.448 0.00124 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.71 on 45 degrees of freedom
## Multiple R-squared: 0.721, Adjusted R-squared: 0.6901
## F-statistic: 23.26 on 5 and 45 DF, p-value: 1.828e-11#The model is statistically significant. With an R square of 72%.